当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nature Communications ( IF 14.7 ) Pub Date : 2018-12-11 , DOI: 10.1038/s41467-018-07722-9 Ravit Netzer 1 , Dina Listov 1 , Rosalie Lipsh 1 , Orly Dym 2 , Shira Albeck 2 , Orli Knop 1 , Colin Kleanthous 3 , Sarel J Fleishman 1
Nature Communications ( IF 14.7 ) Pub Date : 2018-12-11 , DOI: 10.1038/s41467-018-07722-9 Ravit Netzer 1 , Dina Listov 1 , Rosalie Lipsh 1 , Orly Dym 2 , Shira Albeck 2 , Orli Knop 1 , Colin Kleanthous 3 , Sarel J Fleishman 1
Affiliation
|
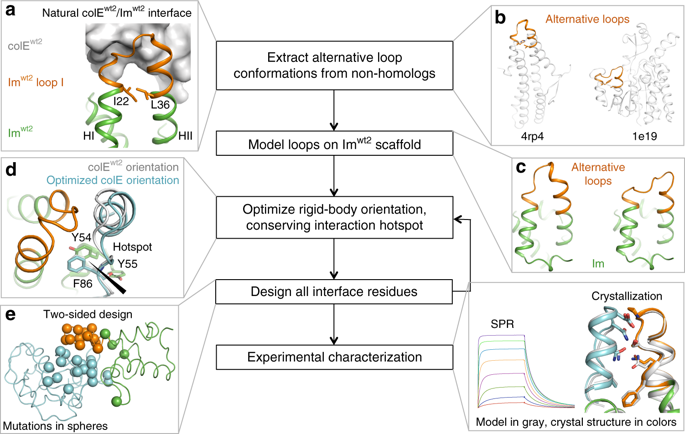
Protein networks in all organisms comprise homologous interacting pairs. In these networks, some proteins are specific, interacting with one or a few binding partners, whereas others are multispecific and bind a range of targets. We describe an algorithm that starts from an interacting pair and designs dozens of new pairs with diverse backbone conformations at the binding site as well as new binding orientations and sequences. Applied to a high-affinity bacterial pair, the algorithm results in 18 new ones, with cognate affinities from pico- to micromolar. Three pairs exhibit 3-5 orders of magnitude switch in specificity relative to the wild type, whereas others are multispecific, collectively forming a protein-interaction network. Crystallographic analysis confirms design accuracy, including in new backbones and polar interactions. Preorganized polar interaction networks are responsible for high specificity, thus defining design principles that can be applied to program synthetic cellular interaction networks of desired affinity and specificity.
中文翻译:

计算设计的蛋白质相互作用对网络中的超高特异性。
所有生物中的蛋白质网络均包含同源相互作用对。在这些网络中,某些蛋白质是特异的,可与一个或几个结合配偶体相互作用,而另一些则是多特异的并结合一系列靶标。我们描述了一种从相互作用对开始的算法,并设计了数十个在结合位点具有不同骨架构象的新对以及新的结合方向和序列。该算法应用于高亲和力细菌对,可产生18个新细菌,关联亲和力从皮摩尔到微摩尔。三对相对于野生型表现出3-5个数量级的特异性转换,而其他对则是多特异性的,共同形成蛋白质相互作用网络。晶体学分析证实了设计的准确性,包括在新的主链和极性相互作用中的准确性。
更新日期:2018-12-11
中文翻译:

计算设计的蛋白质相互作用对网络中的超高特异性。
所有生物中的蛋白质网络均包含同源相互作用对。在这些网络中,某些蛋白质是特异的,可与一个或几个结合配偶体相互作用,而另一些则是多特异的并结合一系列靶标。我们描述了一种从相互作用对开始的算法,并设计了数十个在结合位点具有不同骨架构象的新对以及新的结合方向和序列。该算法应用于高亲和力细菌对,可产生18个新细菌,关联亲和力从皮摩尔到微摩尔。三对相对于野生型表现出3-5个数量级的特异性转换,而其他对则是多特异性的,共同形成蛋白质相互作用网络。晶体学分析证实了设计的准确性,包括在新的主链和极性相互作用中的准确性。
















































 京公网安备 11010802027423号
京公网安备 11010802027423号