当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Single-Nucleotide Resolution Analysis of 5-Hydroxymethylcytosine in DNA by Enzyme-Mediated Deamination in Combination with Sequencing
Analytical Chemistry ( IF 6.7 ) Pub Date : 2018-11-20 00:00:00 , DOI: 10.1021/acs.analchem.8b04833 Qiao-Ying Li 1 , Neng-Bin Xie 1 , Jun Xiong 1 , Bi-Feng Yuan 1 , Yu-Qi Feng 1
Analytical Chemistry ( IF 6.7 ) Pub Date : 2018-11-20 00:00:00 , DOI: 10.1021/acs.analchem.8b04833 Qiao-Ying Li 1 , Neng-Bin Xie 1 , Jun Xiong 1 , Bi-Feng Yuan 1 , Yu-Qi Feng 1
Affiliation
|
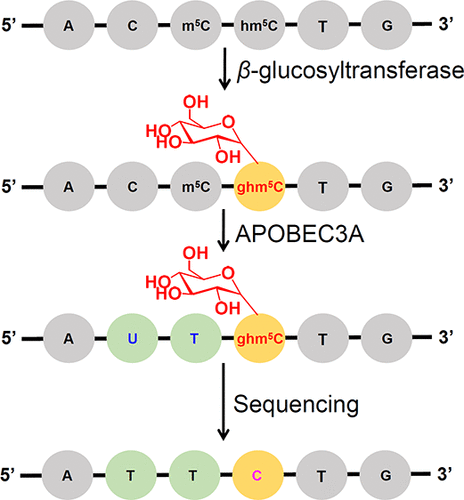
The report of the existence of 5-hydroxymethylcytosine (hm5C) in mammalian genomes is a milestone discovery. hm5C is now generally viewed as the sixth base of DNA with important functions on epigenetic regulation. The in-depth investigation of the biological functions of hm5C requires elucidating the distribution patterns of hm5C in genomes, better in single-nucleotide resolution. It was reported that the cytosine deaminases of the APOBEC (apolipoprotein B mRNA-editing catalytic polypeptide-like) family are nucleic acid editing enzymes and can deaminate cytosine (C) to form uracil (U). Particularly, a subfamily of APOBEC (APOBEC3A) can efficiently deaminate both C and 5-methylcytosine (m5C). In the current study, we identified that APOBEC3A protein can effectively deaminate C, m5C, and hm5C but shows no observable deamination activity toward glycosylated hm5C (β-glucosyl-5-hydroxymethyl-2′-deoxycytidine, ghm5C) by using the restriction enzyme-based assay and liquid chromatography–electrospray ionization–tandem mass spectrometry (LC-ESI-MS/MS) analysis. By virtue of the differential deamination activity of APOBEC3A toward C, m5C, and ghm5C in conjugation with sequencing, we developed the single-nucleotide resolution analysis of hm5C in DNA. In this analytical strategy, the original C and m5C in DNA will be deaminated by APOBEC3A to form U and thymine (T), both of which will read as T during sequencing, while ghm5C is resistant to deamination and will read as C during sequencing. Therefore, the remaining C in the sequence context only could come from original hm5C, which offers the single-nucleotide resolution analysis of hm5C in DNA. This APOBEC3A-mediated deamination sequencing (AMD-seq) is straightforward and involves no bisulfite treatment, which avoids the substantial degradation of DNA. Future application of this strategy can be performed for the reliable mapping of hm5C in genome-wide scale at the single-nucleotide resolution.
中文翻译:

酶介导的脱氨结合测序法测定DNA中5-羟甲基胞嘧啶的单核苷酸分辨率
哺乳动物基因组中存在5-羟甲基胞嘧啶(hm 5 C)的报道是一个里程碑式的发现。hm 5 C现在通常被视为DNA的第六个碱基,在表观遗传调控中具有重要功能。深入研究hm 5 C的生物学功能需要阐明hm 5 C在基因组中的分布模式,单核苷酸分辨率更好。据报道,APOBEC(载脂蛋白B mRNA编辑催化多肽样)家族的胞嘧啶脱氨酶是核酸编辑酶,可以使胞嘧啶(C)脱氨基形成尿嘧啶(U)。特别是,APOBEC的一个亚家族(APOBEC3A)可以有效地将C和5-甲基胞嘧啶脱氨(m 5C)。在当前的研究中,我们发现APOBEC3A蛋白可以有效地使C,m 5 C和hm 5 C脱氨基,但是对糖基化的hm 5 C(β-葡萄糖基-5-羟甲基-2'-脱氧胞苷,ghm 5 C)通过基于限制性酶的分析和液相色谱-电喷雾电离-串联质谱分析(LC-ESI-MS / MS)进行分析。凭借APOBEC3A的差动脱氨酶活性朝向C,M的5 ℃,GHM 5与测序共轭C,我们开发了HM的单核苷酸分辨率分析5中DNA℃。在这种分析策略中,原始的C和m 5DNA中的C将被APOBEC3A脱氨基形成U和胸腺嘧啶(T),两者在测序过程中均读为T,而ghm 5 C具有抗脱氨性,在测序过程中均读为C。因此,序列上下文中的其余C只能来自原始的hm 5 C,它提供了DNA中hm 5 C的单核苷酸分辨率分析。该APOBEC3A介导的脱氨测序(AMD-seq)非常简单,不涉及亚硫酸氢盐处理,从而避免了DNA的大量降解。这个策略的未来的应用程序可以为HM的可靠映射来执行5在全基因组规模的单核苷酸分辨率℃。
更新日期:2018-11-20
中文翻译:

酶介导的脱氨结合测序法测定DNA中5-羟甲基胞嘧啶的单核苷酸分辨率
哺乳动物基因组中存在5-羟甲基胞嘧啶(hm 5 C)的报道是一个里程碑式的发现。hm 5 C现在通常被视为DNA的第六个碱基,在表观遗传调控中具有重要功能。深入研究hm 5 C的生物学功能需要阐明hm 5 C在基因组中的分布模式,单核苷酸分辨率更好。据报道,APOBEC(载脂蛋白B mRNA编辑催化多肽样)家族的胞嘧啶脱氨酶是核酸编辑酶,可以使胞嘧啶(C)脱氨基形成尿嘧啶(U)。特别是,APOBEC的一个亚家族(APOBEC3A)可以有效地将C和5-甲基胞嘧啶脱氨(m 5C)。在当前的研究中,我们发现APOBEC3A蛋白可以有效地使C,m 5 C和hm 5 C脱氨基,但是对糖基化的hm 5 C(β-葡萄糖基-5-羟甲基-2'-脱氧胞苷,ghm 5 C)通过基于限制性酶的分析和液相色谱-电喷雾电离-串联质谱分析(LC-ESI-MS / MS)进行分析。凭借APOBEC3A的差动脱氨酶活性朝向C,M的5 ℃,GHM 5与测序共轭C,我们开发了HM的单核苷酸分辨率分析5中DNA℃。在这种分析策略中,原始的C和m 5DNA中的C将被APOBEC3A脱氨基形成U和胸腺嘧啶(T),两者在测序过程中均读为T,而ghm 5 C具有抗脱氨性,在测序过程中均读为C。因此,序列上下文中的其余C只能来自原始的hm 5 C,它提供了DNA中hm 5 C的单核苷酸分辨率分析。该APOBEC3A介导的脱氨测序(AMD-seq)非常简单,不涉及亚硫酸氢盐处理,从而避免了DNA的大量降解。这个策略的未来的应用程序可以为HM的可靠映射来执行5在全基因组规模的单核苷酸分辨率℃。































 京公网安备 11010802027423号
京公网安备 11010802027423号