The EMBO Journal ( IF 9.4 ) Pub Date : 2018-11-02 , DOI: 10.15252/embj.201899278 Marine Dehecq 1, 2 , Laurence Decourty 1 , Abdelkader Namane 1 , Caroline Proux 3 , Joanne Kanaan 4 , Hervé Le Hir 4 , Alain Jacquier 1 , Cosmin Saveanu 1
|
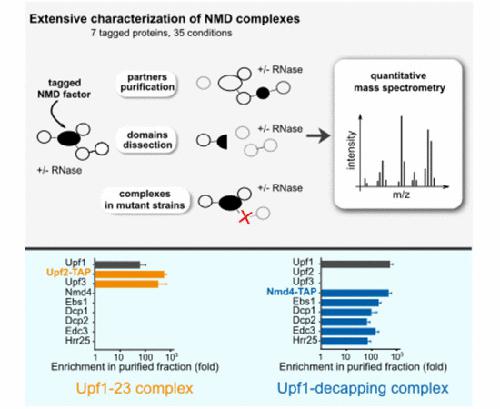
Nonsense‐mediated mRNA decay (NMD) is a translation‐dependent RNA degradation pathway involved in many cellular pathways and crucial for telomere maintenance and embryo development. Core NMD factors Upf1, Upf2 and Upf3 are conserved from yeast to mammals, but a universal NMD model is lacking. We used affinity purification coupled with mass spectrometry and an improved data analysis protocol to characterize the composition and dynamics of yeast NMD complexes in yeast (112 experiments). Unexpectedly, we identified two distinct complexes associated with Upf1: Upf1‐23 (Upf1, Upf2, Upf3) and Upf1‐decapping. Upf1‐decapping contained the mRNA decapping enzyme, together with Nmd4 and Ebs1, two proteins that globally affected NMD and were critical for RNA degradation mediated by the Upf1 C‐terminal helicase region. The fact that Nmd4 association with RNA was partially dependent on Upf1‐23 components and the similarity between Nmd4/Ebs1 and mammalian Smg5‐7 proteins suggest that NMD operates through conserved, successive Upf1‐23 and Upf1‐decapping complexes. This model can be extended to accommodate steps that are missing in yeast, to serve for further mechanistic studies of NMD in eukaryotes.
中文翻译:

无义介导的mRNA衰变涉及两个截然不同的Upf1结合复合体
无义介导的mRNA衰变(NMD)是翻译依赖的RNA降解途径,涉及许多细胞途径,对于端粒的维持和胚胎发育至关重要。核心NMD因子Upf1,Upf2和Upf3从酵母到哺乳动物都是保守的,但缺乏通用的NMD模型。我们使用亲和纯化结合质谱和改进的数据分析协议来表征酵母中酵母NMD复合物的组成和动力学(112个实验)。出乎意料的是,我们发现与Upf1的相关的两个不同的复合物:Upf1-23(Upf1的,UPF2,UPF3)和Upf1的,开模。Upf1上限包含mRNA脱盖酶,以及Nmd4和Ebs1,这两种蛋白在全球范围内影响NMD,并且对Upf1 C端解旋酶区域介导的RNA降解至关重要。Nmd4与RNA的结合部分取决于Upf1-23成分,并且Nmd4 / Ebs1与哺乳动物Smg5-7蛋白之间的相似性表明NMD通过保守的,连续的Upf1-23和Upf1脱盖复合物起作用。可以扩展该模型以适应酵母中缺少的步骤,从而为真核生物中NMD的进一步机理研究服务。

































 京公网安备 11010802027423号
京公网安备 11010802027423号