Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 α-1,3-glucanase from Paenibacillus glycanilyticus FH11.
The FEBS Journal ( IF 5.5 ) Pub Date : 2019-12-02 , DOI: 10.1111/febs.15161 Takafumi Itoh 1 , Rattanaporn Intuy 2 , Wasana Suyotha 3 , Junji Hayashi 2 , Shigekazu Yano 4 , Koki Makabe 4 , Mamoru Wakayama 2 , Takao Hibi 1
The FEBS Journal ( IF 5.5 ) Pub Date : 2019-12-02 , DOI: 10.1111/febs.15161 Takafumi Itoh 1 , Rattanaporn Intuy 2 , Wasana Suyotha 3 , Junji Hayashi 2 , Shigekazu Yano 4 , Koki Makabe 4 , Mamoru Wakayama 2 , Takao Hibi 1
Affiliation
|
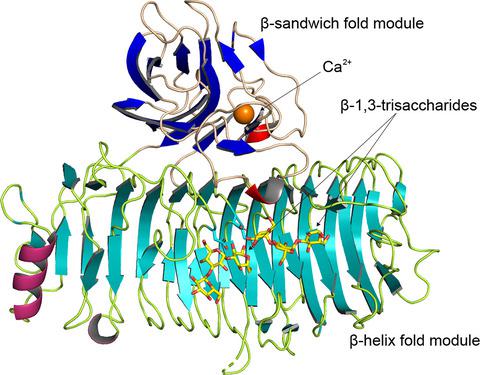
The α‐1,3‐glucanase from Paenibacillus glycanilyticus FH11 (Agl‐FH1), a member of the glycoside hydrolase family 87 (GH87), hydrolyzes α‐1,3‐glucan with an endo ‐action. GH87 enzymes are known to degrade dental plaque produced by oral pathogenic Streptococcus species. In this study, the kinetic analyses revealed that this enzyme hydrolyzed α‐1,3‐tetraglucan into glucose and α‐1,3‐triglucan with β‐configuration at the reducing end by an inverting mechanism. The crystal structures of the catalytic domain (CatAgl‐FH1) complexed with or without oligosaccharides at 1.4–2.5 or 1.6 Å resolutions, respectively, are also presented. The initial crystal structure of CatAgl‐FH1 was determined by native single‐wavelength anomalous diffraction. The catalytic domain was composed of two modules, a β‐sandwich fold module, and a right‐handed β‐helix fold module. The structure of the β‐sandwich was similar to those of the carbohydrate‐binding module family 35 members. The glycerol or nigerose enzyme complex structures demonstrated that this β‐sandwich fold module is a novel carbohydrate‐binding module with the capabilities to bind saccharides and to promote the degradation of polysaccharides. The structures of the inactive mutant in complexes with oligosaccharide showed that at least eight subsites for glucose binding were located in the active cleft of the β‐helix fold and the architecture of the active cleft was suitable for the recognition and hydrolysis of α‐1,3‐glucan by the inverting mechanism. The structural similarity to GH28 and GH49 enzymes and the results of site‐directed mutagenesis indicated that three Asp residues, Asp1045, Asp1068, and Asp1069, are the most likely candidates for the catalytic residues of Agl‐FH1.
中文翻译:

从糖胺酸芽孢杆菌FH11的糖苷水解酶家族87α-1,3-葡聚糖酶对底物识别和催化的结构见解。
糖化水解酶家族87(GH87)的成员糖胺酸杆状芽孢杆菌FH11(Ag1-FH1)的α-1,3-葡聚糖酶可水解内聚作用的α-1,3-葡聚糖。已知GH87酶可降解口腔致病性链球菌产生的牙菌斑物种。在这项研究中,动力学分析表明,该酶通过转化机制将α-1,3-四葡聚糖水解为葡萄糖和还原端具有β-构型的α-1,3-三葡聚糖。还介绍了催化结构域(CatAg1-FH1)的晶体结构,有或没有寡糖分别以1.4–2.5或1.6Å的分辨率结合。CatAg1-FH1的初始晶体结构是通过自然单波长异常衍射确定的。催化域由两个模块组成,一个β夹心折叠模块和一个右手β螺旋折叠模块。β三明治的结构与糖结合模块家族35个成员的结构相似。甘油或黑糖酶复合物结构表明,这种β-三明治折叠模块是一种新型的碳水化合物结合模块,具有结合糖类并促进多糖降解的能力。与寡糖复合的无活性突变体的结构表明,至少有八个与葡萄糖结合的亚位点位于β-螺旋折叠的活性裂隙中,并且该活性裂隙的结构适合于α-1的识别和水解, 3-葡聚糖的反相机制。与GH28和GH49酶的结构相似性以及定点诱变的结果表明,三个Asp残基Asp1045,Asp1068和Asp1069最可能是Ag1-FH1催化残基的候选对象。与寡糖复合的无活性突变体的结构表明,至少有八个与葡萄糖结合的亚位点位于β-螺旋折叠的活性裂隙中,并且该活性裂隙的结构适合于α-1的识别和水解, 3-葡聚糖的反相机制。与GH28和GH49酶的结构相似性以及定点诱变的结果表明,三个Asp残基Asp1045,Asp1068和Asp1069最可能是Ag1-FH1催化残基的候选对象。与寡糖复合的无活性突变体的结构表明,至少有八个与葡萄糖结合的亚位点位于β-螺旋折叠的活性裂隙中,并且该活性裂隙的结构适合于α-1的识别和水解, 3-葡聚糖的反相机制。与GH28和GH49酶的结构相似性以及定点诱变的结果表明,三个Asp残基Asp1045,Asp1068和Asp1069最可能是Ag1-FH1催化残基的候选对象。
更新日期:2019-12-02
中文翻译:

从糖胺酸芽孢杆菌FH11的糖苷水解酶家族87α-1,3-葡聚糖酶对底物识别和催化的结构见解。
糖化水解酶家族87(GH87)的成员糖胺酸杆状芽孢杆菌FH11(Ag1-FH1)的α-1,3-葡聚糖酶可水解内聚作用的α-1,3-葡聚糖。已知GH87酶可降解口腔致病性链球菌产生的牙菌斑物种。在这项研究中,动力学分析表明,该酶通过转化机制将α-1,3-四葡聚糖水解为葡萄糖和还原端具有β-构型的α-1,3-三葡聚糖。还介绍了催化结构域(CatAg1-FH1)的晶体结构,有或没有寡糖分别以1.4–2.5或1.6Å的分辨率结合。CatAg1-FH1的初始晶体结构是通过自然单波长异常衍射确定的。催化域由两个模块组成,一个β夹心折叠模块和一个右手β螺旋折叠模块。β三明治的结构与糖结合模块家族35个成员的结构相似。甘油或黑糖酶复合物结构表明,这种β-三明治折叠模块是一种新型的碳水化合物结合模块,具有结合糖类并促进多糖降解的能力。与寡糖复合的无活性突变体的结构表明,至少有八个与葡萄糖结合的亚位点位于β-螺旋折叠的活性裂隙中,并且该活性裂隙的结构适合于α-1的识别和水解, 3-葡聚糖的反相机制。与GH28和GH49酶的结构相似性以及定点诱变的结果表明,三个Asp残基Asp1045,Asp1068和Asp1069最可能是Ag1-FH1催化残基的候选对象。与寡糖复合的无活性突变体的结构表明,至少有八个与葡萄糖结合的亚位点位于β-螺旋折叠的活性裂隙中,并且该活性裂隙的结构适合于α-1的识别和水解, 3-葡聚糖的反相机制。与GH28和GH49酶的结构相似性以及定点诱变的结果表明,三个Asp残基Asp1045,Asp1068和Asp1069最可能是Ag1-FH1催化残基的候选对象。与寡糖复合的无活性突变体的结构表明,至少有八个与葡萄糖结合的亚位点位于β-螺旋折叠的活性裂隙中,并且该活性裂隙的结构适合于α-1的识别和水解, 3-葡聚糖的反相机制。与GH28和GH49酶的结构相似性以及定点诱变的结果表明,三个Asp残基Asp1045,Asp1068和Asp1069最可能是Ag1-FH1催化残基的候选对象。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号