当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Automated sequence design of 2D wireframe DNA origami with honeycomb edges.
Nature Communications ( IF 14.7 ) Pub Date : 2019-11-28 , DOI: 10.1038/s41467-019-13457-y Hyungmin Jun 1 , Xiao Wang 1 , William P Bricker 1 , Mark Bathe 1
Nature Communications ( IF 14.7 ) Pub Date : 2019-11-28 , DOI: 10.1038/s41467-019-13457-y Hyungmin Jun 1 , Xiao Wang 1 , William P Bricker 1 , Mark Bathe 1
Affiliation
|
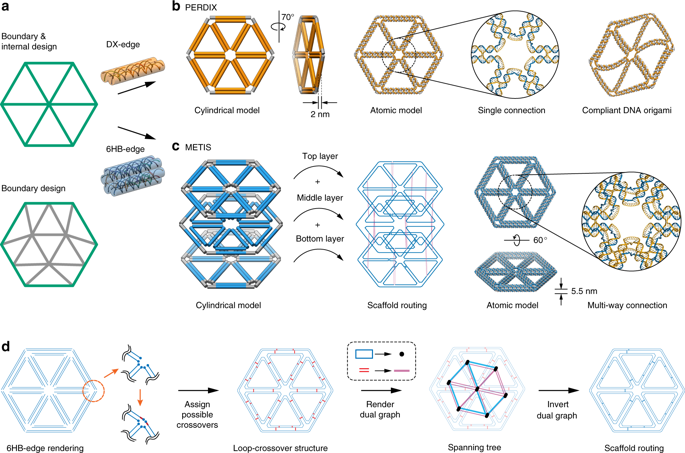
Wireframe DNA origami has emerged as a powerful approach to fabricating nearly arbitrary 2D and 3D geometries at the nanometer-scale. Complex scaffold and staple routing needed to design wireframe DNA origami objects, however, render fully automated, geometry-based sequence design approaches essential for their synthesis. And wireframe DNA origami structural fidelity can be limited by wireframe edges that are composed only of one or two duplexes. Here we introduce a fully automated computational approach that programs 2D wireframe origami assemblies using honeycomb edges composed of six parallel duplexes. These wireframe assemblies show enhanced structural fidelity from electron microscopy-based measurement of programmed angles compared with identical geometries programmed using dual-duplex edges. Molecular dynamics provides additional theoretical support for the enhanced structural fidelity observed. Application of our top-down sequence design procedure to a variety of complex objects demonstrates its broad utility for programmable 2D nanoscale materials.
中文翻译:

具有蜂窝状边缘的2D线框DNA折纸的自动序列设计。
线框DNA折纸已成为一种强大的方法,可以在纳米级上制造几乎任意的2D和3D几何形状。设计线框DNA折纸对象需要复杂的脚手架和订书钉布线,但是,它们提供了全自动,基于几何的序列设计方法,对于它们的合成至关重要。线框DNA折纸的结构保真度可能受到仅由一个或两个双链体组成的线框边缘的限制。在这里,我们介绍一种全自动的计算方法,该方法使用由六个平行双工组成的蜂窝状边缘对2D线框折纸装配进行编程。与使用双双工边编程的相同几何形状相比,这些线框组件通过基于电子显微镜的编程角度测量显示出更高的结构保真度。分子动力学为观察到的增强的结构保真度提供了额外的理论支持。将我们的自上而下的序列设计程序应用于各种复杂对象,证明了其在可编程2D纳米级材料中的广泛用途。
更新日期:2019-11-29
中文翻译:

具有蜂窝状边缘的2D线框DNA折纸的自动序列设计。
线框DNA折纸已成为一种强大的方法,可以在纳米级上制造几乎任意的2D和3D几何形状。设计线框DNA折纸对象需要复杂的脚手架和订书钉布线,但是,它们提供了全自动,基于几何的序列设计方法,对于它们的合成至关重要。线框DNA折纸的结构保真度可能受到仅由一个或两个双链体组成的线框边缘的限制。在这里,我们介绍一种全自动的计算方法,该方法使用由六个平行双工组成的蜂窝状边缘对2D线框折纸装配进行编程。与使用双双工边编程的相同几何形状相比,这些线框组件通过基于电子显微镜的编程角度测量显示出更高的结构保真度。分子动力学为观察到的增强的结构保真度提供了额外的理论支持。将我们的自上而下的序列设计程序应用于各种复杂对象,证明了其在可编程2D纳米级材料中的广泛用途。































 京公网安备 11010802027423号
京公网安备 11010802027423号