Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 Systems.
Molecular Cell ( IF 14.5 ) Pub Date : 2019-11-19 , DOI: 10.1016/j.molcel.2019.10.024 Liang-Zhong Yang 1 , Yang Wang 1 , Si-Qi Li 1 , Run-Wen Yao 1 , Peng-Fei Luan 1 , Huang Wu 1 , Gordon G Carmichael 2 , Ling-Ling Chen 3
Molecular Cell ( IF 14.5 ) Pub Date : 2019-11-19 , DOI: 10.1016/j.molcel.2019.10.024 Liang-Zhong Yang 1 , Yang Wang 1 , Si-Qi Li 1 , Run-Wen Yao 1 , Peng-Fei Luan 1 , Huang Wu 1 , Gordon G Carmichael 2 , Ling-Ling Chen 3
Affiliation
|
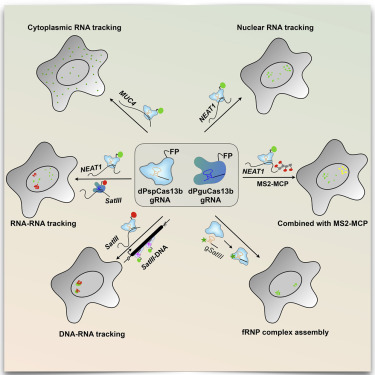
Visualizing the location and dynamics of RNAs in live cells is key to understanding their function. Here, we identify two endonuclease-deficient, single-component programmable RNA-guided and RNA-targeting Cas13 RNases (dCas13s) that allow robust real-time imaging and tracking of RNAs in live cells, even when using single 20- to 27-nt-long guide RNAs. Compared to the aptamer-based MS2-MCP strategy, an optimized dCas13 system is user friendly, does not require genetic manipulation, and achieves comparable RNA-labeling efficiency. We demonstrate that the dCas13 system is capable of labeling NEAT1, SatIII, MUC4, and GCN4 RNAs and allows the study of paraspeckle-associated NEAT1 dynamics. Applying orthogonal dCas13 proteins or combining dCas13 and MS2-MCP allows dual-color imaging of RNAs in single cells. Further combination of dCas13 and dCas9 systems allows simultaneous visualization of genomic DNA and RNA transcripts in living cells.
中文翻译:

通过 CRISPR-Cas13 系统对活细胞中的 RNA 进行动态成像。
可视化活细胞中 RNA 的位置和动态是了解其功能的关键。在这里,我们鉴定了两种核酸内切酶缺陷型、单组分可编程 RNA 引导和 RNA 靶向 Cas13 RNase (dCas13),即使在使用单个 20 至 27 nt 的情况下,也可以对活细胞中的 RNA 进行可靠的实时成像和跟踪-长向导RNA。与基于适体的 MS2-MCP 策略相比,优化的 dCas13 系统用户友好,不需要基因操作,并实现了可比的 RNA 标记效率。我们证明 dCas13 系统能够标记 NEAT1、SatIII、MUC4 和 GCN4 RNA,并允许研究 paraspeckle 相关的 NEAT1 动力学。应用正交 dCas13 蛋白或组合 dCas13 和 MS2-MCP 可以对单细胞中的 RNA 进行双色成像。 dCas13 和 dCas9 系统的进一步组合允许同时可视化活细胞中的基因组 DNA 和 RNA 转录本。
更新日期:2019-11-20
中文翻译:

通过 CRISPR-Cas13 系统对活细胞中的 RNA 进行动态成像。
可视化活细胞中 RNA 的位置和动态是了解其功能的关键。在这里,我们鉴定了两种核酸内切酶缺陷型、单组分可编程 RNA 引导和 RNA 靶向 Cas13 RNase (dCas13),即使在使用单个 20 至 27 nt 的情况下,也可以对活细胞中的 RNA 进行可靠的实时成像和跟踪-长向导RNA。与基于适体的 MS2-MCP 策略相比,优化的 dCas13 系统用户友好,不需要基因操作,并实现了可比的 RNA 标记效率。我们证明 dCas13 系统能够标记 NEAT1、SatIII、MUC4 和 GCN4 RNA,并允许研究 paraspeckle 相关的 NEAT1 动力学。应用正交 dCas13 蛋白或组合 dCas13 和 MS2-MCP 可以对单细胞中的 RNA 进行双色成像。 dCas13 和 dCas9 系统的进一步组合允许同时可视化活细胞中的基因组 DNA 和 RNA 转录本。































 京公网安备 11010802027423号
京公网安备 11010802027423号