Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Locally Fixed Alleles: A method to localize gene drive to island populations.
Scientific Reports ( IF 3.8 ) Pub Date : 2019-11-01 , DOI: 10.1038/s41598-019-51994-0 Jaye Sudweeks 1 , Brandon Hollingsworth 2 , Dimitri V Blondel 3 , Karl J Campbell 4 , Sumit Dhole 5 , John D Eisemann 6 , Owain Edwards 7 , John Godwin 3, 8 , Gregg R Howald 4 , Kevin P Oh 6, 9 , Antoinette J Piaggio 6 , Thomas A A Prowse 10 , Joshua V Ross 10 , J Royden Saah 4, 8 , Aaron B Shiels 6 , Paul Q Thomas 11 , David W Threadgill 12 , Michael R Vella 2 , Fred Gould 5, 8 , Alun L Lloyd 1, 2
Scientific Reports ( IF 3.8 ) Pub Date : 2019-11-01 , DOI: 10.1038/s41598-019-51994-0 Jaye Sudweeks 1 , Brandon Hollingsworth 2 , Dimitri V Blondel 3 , Karl J Campbell 4 , Sumit Dhole 5 , John D Eisemann 6 , Owain Edwards 7 , John Godwin 3, 8 , Gregg R Howald 4 , Kevin P Oh 6, 9 , Antoinette J Piaggio 6 , Thomas A A Prowse 10 , Joshua V Ross 10 , J Royden Saah 4, 8 , Aaron B Shiels 6 , Paul Q Thomas 11 , David W Threadgill 12 , Michael R Vella 2 , Fred Gould 5, 8 , Alun L Lloyd 1, 2
Affiliation
|
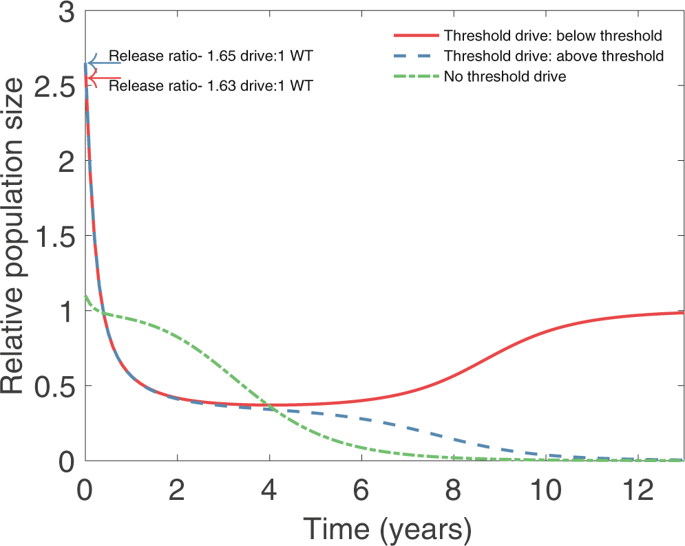
Invasive species pose a major threat to biodiversity on islands. While successes have been achieved using traditional removal methods, such as toxicants aimed at rodents, these approaches have limitations and various off-target effects on island ecosystems. Gene drive technologies designed to eliminate a population provide an alternative approach, but the potential for drive-bearing individuals to escape from the target release area and impact populations elsewhere is a major concern. Here we propose the "Locally Fixed Alleles" approach as a novel means for localizing elimination by a drive to an island population that exhibits significant genetic isolation from neighboring populations. Our approach is based on the assumption that in small island populations of rodents, genetic drift will lead to alleles at multiple genomic loci becoming fixed. In contrast, multiple alleles are likely to be maintained in larger populations on mainlands. Utilizing the high degree of genetic specificity achievable using homing drives, for example based on the CRISPR/Cas9 system, our approach aims at employing one or more locally fixed alleles as the target for a gene drive on a particular island. Using mathematical modeling, we explore the feasibility of this approach and the degree of localization that can be achieved. We show that across a wide range of parameter values, escape of the drive to a neighboring population in which the target allele is not fixed will at most lead to modest transient suppression of the non-target population. While the main focus of this paper is on elimination of a rodent pest from an island, we also discuss the utility of the locally fixed allele approach for the goals of population suppression or population replacement. Our analysis also provides a threshold condition for the ability of a gene drive to invade a partially resistant population.
中文翻译:

局部固定的等位基因:一种将基因驱动定位到岛上种群的方法。
外来入侵物种对岛屿上的生物多样性构成了重大威胁。尽管使用传统的清除方法(例如针对啮齿动物的有毒物质)取得了成功,但这些方法仍存在局限性,并且对岛屿生态系统产生各种脱靶效应。旨在消除种群的基因驱动技术提供了另一种方法,但具有驱动力的个体逃脱目标释放区域并影响其他地方种群的潜力是一个主要问题。在这里,我们提出“局部固定的等位基因”方法,作为一种通过驱赶到与邻近种群表现出显着遗传隔离的岛屿种群来进行局部消灭的新颖手段。我们的方法基于以下假设:在小岛上的啮齿动物中,遗传漂移将导致多个基因组位点的等位基因变得固定。相比之下,大陆的较大人群中可能会保留多个等位基因。利用例如基于CRISPR / Cas9系统的归巢驱动器可以实现的高度遗传特异性,我们的方法旨在采用一个或多个局部固定的等位基因作为特定岛上基因驱动器的目标。使用数学建模,我们探索了这种方法的可行性以及可以实现的本地化程度。我们表明,在广泛的参数值范围内,驱动器逃逸至目标等位基因未固定的邻近种群最多将导致对非目标种群的适度瞬态抑制。尽管本文的主要重点是消除岛屿上的啮齿动物害虫,我们还讨论了局部固定等位基因方法在抑制种群或替代种群中的作用。我们的分析还为基因驱动力入侵部分抗性种群的能力提供了一个阈值条件。
更新日期:2019-11-01
中文翻译:

局部固定的等位基因:一种将基因驱动定位到岛上种群的方法。
外来入侵物种对岛屿上的生物多样性构成了重大威胁。尽管使用传统的清除方法(例如针对啮齿动物的有毒物质)取得了成功,但这些方法仍存在局限性,并且对岛屿生态系统产生各种脱靶效应。旨在消除种群的基因驱动技术提供了另一种方法,但具有驱动力的个体逃脱目标释放区域并影响其他地方种群的潜力是一个主要问题。在这里,我们提出“局部固定的等位基因”方法,作为一种通过驱赶到与邻近种群表现出显着遗传隔离的岛屿种群来进行局部消灭的新颖手段。我们的方法基于以下假设:在小岛上的啮齿动物中,遗传漂移将导致多个基因组位点的等位基因变得固定。相比之下,大陆的较大人群中可能会保留多个等位基因。利用例如基于CRISPR / Cas9系统的归巢驱动器可以实现的高度遗传特异性,我们的方法旨在采用一个或多个局部固定的等位基因作为特定岛上基因驱动器的目标。使用数学建模,我们探索了这种方法的可行性以及可以实现的本地化程度。我们表明,在广泛的参数值范围内,驱动器逃逸至目标等位基因未固定的邻近种群最多将导致对非目标种群的适度瞬态抑制。尽管本文的主要重点是消除岛屿上的啮齿动物害虫,我们还讨论了局部固定等位基因方法在抑制种群或替代种群中的作用。我们的分析还为基因驱动力入侵部分抗性种群的能力提供了一个阈值条件。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号