当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions.
Nature Communications ( IF 14.7 ) Pub Date : 2015-Nov-09 , DOI: 10.1038/ncomms9720 Andrew D. Doyle , Nicole Carvajal , Albert Jin , Kazue Matsumoto , Kenneth M. Yamada
Nature Communications ( IF 14.7 ) Pub Date : 2015-Nov-09 , DOI: 10.1038/ncomms9720 Andrew D. Doyle , Nicole Carvajal , Albert Jin , Kazue Matsumoto , Kenneth M. Yamada
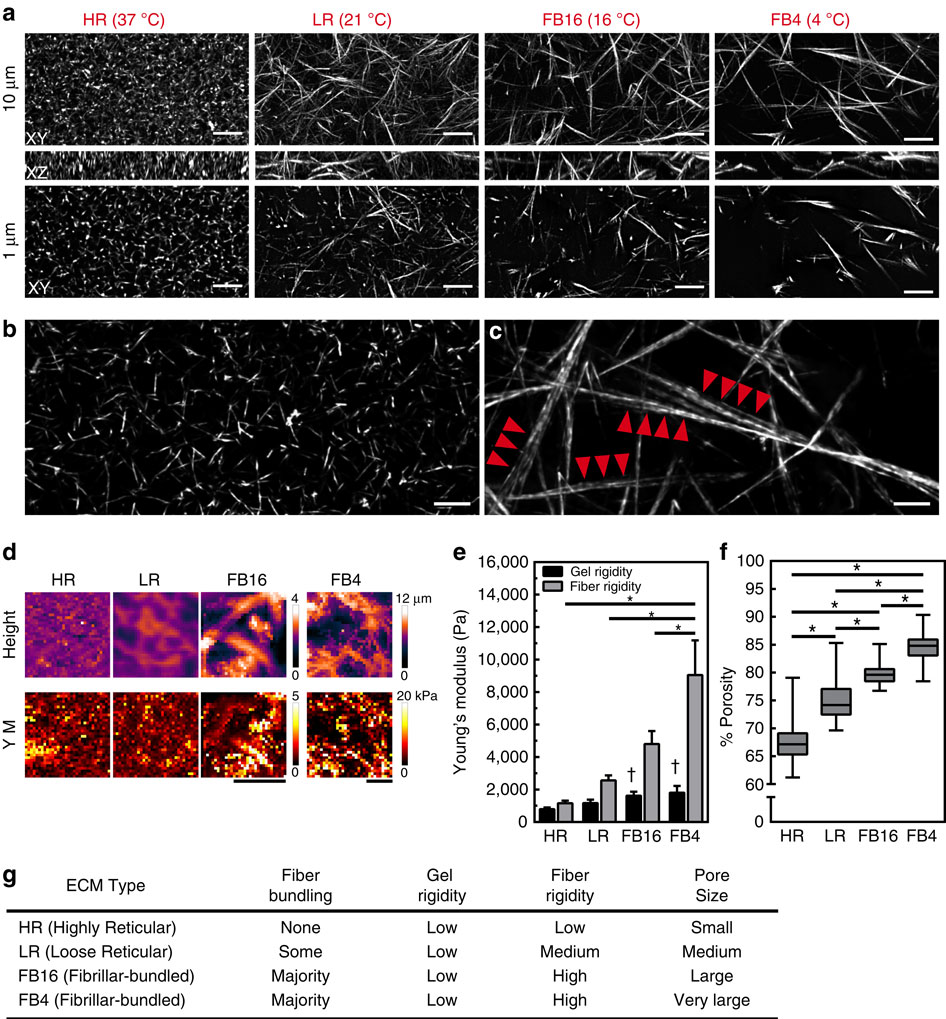
|
The physical properties of two-dimensional (2D) extracellular matrices (ECMs) modulate cell adhesion dynamics and motility, but little is known about the roles of local microenvironmental differences in three-dimensional (3D) ECMs. Here we generate 3D collagen gels of varying matrix microarchitectures to characterize their regulation of 3D adhesion dynamics and cell migration. ECMs containing bundled fibrils demonstrate enhanced local adhesion-scale stiffness and increased adhesion stability through balanced ECM/adhesion coupling, whereas highly pliable reticular matrices promote adhesion retraction. 3D adhesion dynamics are locally regulated by ECM rigidity together with integrin/ECM association and myosin II contractility. Unlike 2D migration, abrogating contractility stalls 3D migration regardless of ECM pore size. We find force is not required for clustering of activated integrins on 3D native collagen fibrils. We propose that efficient 3D migration requires local balancing of contractility with ECM stiffness to stabilize adhesions, which facilitates the detachment of activated integrins from ECM fibrils.
中文翻译:

局部3D矩阵微环境通过时空动力学的收缩依赖性黏附调节细胞迁移。
二维(2D)细胞外基质(ECM)的物理特性调节细胞粘附动力学和运动性,但对三维(3D)ECM中局部微环境差异的作用了解甚少。在这里,我们生成了具有不同基质微结构的3D胶原蛋白凝胶,以表征其对3D粘附动力学和细胞迁移的调节。包含成束原纤维的ECM通过平衡的ECM /黏附耦合表现出增强的局部黏附规模刚度和增加的黏附稳定性,而高度柔韧的网状基质则促进黏附收缩。3D粘附动力学受ECM刚性,整联蛋白/ ECM缔合和肌球蛋白II收缩力的调节。与2D迁移不同,无论ECM孔径如何,取消收缩都将阻止3D迁移。我们发现在3D天然胶原蛋白原纤维上活化的整联蛋白的簇集不需要力。我们提出有效的3D迁移需要在收缩力和ECM刚度之间保持局部平衡,以稳定粘连,这有助于将活化的整联蛋白从ECM原纤维中分离出来。
更新日期:2015-11-12
中文翻译:

局部3D矩阵微环境通过时空动力学的收缩依赖性黏附调节细胞迁移。
二维(2D)细胞外基质(ECM)的物理特性调节细胞粘附动力学和运动性,但对三维(3D)ECM中局部微环境差异的作用了解甚少。在这里,我们生成了具有不同基质微结构的3D胶原蛋白凝胶,以表征其对3D粘附动力学和细胞迁移的调节。包含成束原纤维的ECM通过平衡的ECM /黏附耦合表现出增强的局部黏附规模刚度和增加的黏附稳定性,而高度柔韧的网状基质则促进黏附收缩。3D粘附动力学受ECM刚性,整联蛋白/ ECM缔合和肌球蛋白II收缩力的调节。与2D迁移不同,无论ECM孔径如何,取消收缩都将阻止3D迁移。我们发现在3D天然胶原蛋白原纤维上活化的整联蛋白的簇集不需要力。我们提出有效的3D迁移需要在收缩力和ECM刚度之间保持局部平衡,以稳定粘连,这有助于将活化的整联蛋白从ECM原纤维中分离出来。

































 京公网安备 11010802027423号
京公网安备 11010802027423号