Biochimica et Biophysica Acta (BBA) - General Subjects ( IF 2.8 ) Pub Date : 2019-03-06 , DOI: 10.1016/j.bbagen.2019.03.003
Ryoji Miyazaki 1 , Yoshinori Akiyama 1 , Hiroyuki Mori 1
|
Background
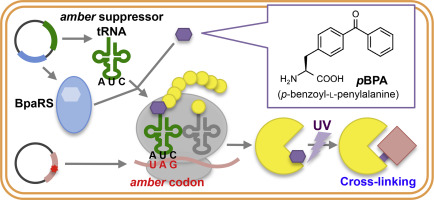
Proteins, which comprise one of the major classes of biomolecules that constitute a cell, interact with other cellular factors during both their biogenesis and functional states. Studying not only static but also transient interactions of proteins is important to understand their physiological roles and regulation mechanisms. However, only a limited number of methods are available to analyze the dynamic behaviors of proteins at the molecular level in a living cell. The site-directed in vivo photo-cross-linking approach is an elegant technique to capture protein interactions with high spatial resolution in a living cell.
Scope of review
Here, we review the in vivo photo-cross-linking approach including its recent applications and the potential problems to be considered. We also introduce a new in vivo photo-cross-linking-based technique (PiXie) to study protein dynamics with high spatiotemporal resolution.
Major conclusions
In vivo photo-cross-linking enables us to capture weak/transient protein interactions with high spatial resolution, and allows for identification of interacting factors. Moreover, the PiXie approach can be used to monitor rapid folding/assembly processes of proteins in living cells.
General significance
In vivo photo-cross-linking is a simple method that has been used to analyze the dynamic interactions of many cellular proteins. Originally developed in Escherichia coli, this system has been extended to studies in various organisms, making it a fundamental technique for investigating dynamic protein interactions in many cellular processes. This article is part of a Special issue entitled “Novel major techniques for visualizing ‘live’ protein molecules” edited by Dr. Daisuke Kohda.
中文翻译:

一种光交联方法来监测活细胞中的蛋白质动态。
背景
蛋白质是构成细胞的主要生物分子之一,它们在其生物发生和功能状态期间都与其他细胞因子相互作用。不仅要研究蛋白质的静态相互作用,还要研究蛋白质的瞬时相互作用对于理解其生理作用和调节机制非常重要。但是,只有有限数量的方法可用于在活细胞中分析分子水平上蛋白质的动态行为。定点体内光交联方法是一种捕获活细胞中具有高空间分辨率的蛋白质相互作用的优雅技术。
审查范围
在此,我们回顾了体内光交联方法,包括其最近的应用以及需要考虑的潜在问题。我们还介绍了一种新的基于体内光交联的技术(PiXie),以高时空分辨率研究蛋白质动力学。
主要结论
体内光交联使我们能够以高空间分辨率捕获弱/瞬态蛋白质相互作用,并能够识别相互作用因子。此外,PiXie方法可用于监测活细胞中蛋白质的快速折叠/组装过程。
一般意义
体内光交联是一种简单的方法,已用于分析许多细胞蛋白的动态相互作用。该系统最初在大肠杆菌中开发,现已扩展到各种生物的研究,使其成为研究许多细胞过程中动态蛋白质相互作用的基本技术。本文是由Daisuke Kohda博士编辑的题为“新颖的可视化“活”蛋白分子的主要技术”的特刊的一部分。































 京公网安备 11010802027423号
京公网安备 11010802027423号