当前位置:
X-MOL 学术
›
Nat. Protoc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integrating hydrogen-deuterium exchange mass spectrometry with molecular dynamics simulations to probe lipid-modulated conformational changes in membrane proteins.
Nature Protocols ( IF 13.1 ) Pub Date : 2019-10-11 , DOI: 10.1038/s41596-019-0219-6 Chloe Martens 1, 2 , Mrinal Shekhar 3 , Andy M Lau 1 , Emad Tajkhorshid 3 , Argyris Politis 1
Nature Protocols ( IF 13.1 ) Pub Date : 2019-10-11 , DOI: 10.1038/s41596-019-0219-6 Chloe Martens 1, 2 , Mrinal Shekhar 3 , Andy M Lau 1 , Emad Tajkhorshid 3 , Argyris Politis 1
Affiliation
|
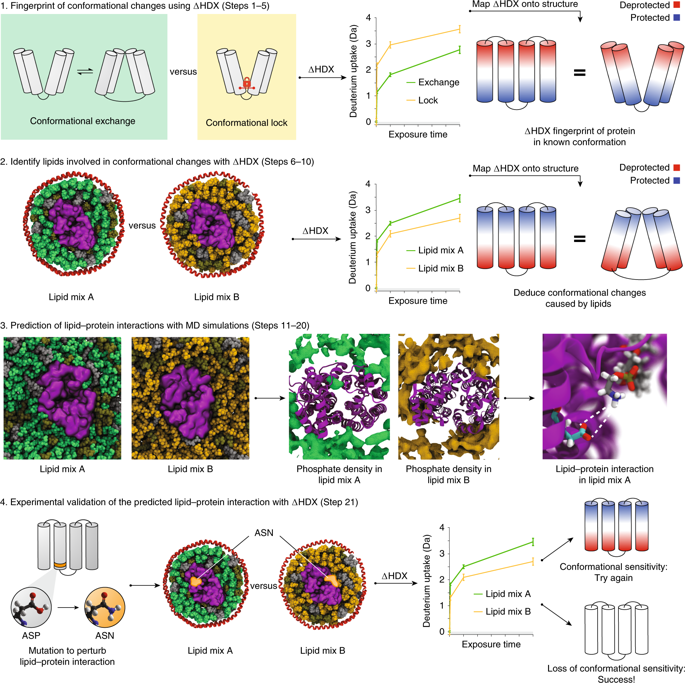
Biological membranes define the boundaries of cells and are composed primarily of phospholipids and membrane proteins. It has become increasingly evident that direct interactions of membrane proteins with their surrounding lipids play key roles in regulating both protein conformations and function. However, the exact nature and structural consequences of these interactions remain difficult to track at the molecular level. Here, we present a protocol that specifically addresses this challenge. First, hydrogen-deuterium exchange mass spectrometry (HDX-MS) of membrane proteins incorporated into nanodiscs of controlled lipid composition is used to obtain information on the lipid species that are involved in modulating the conformational changes in the membrane protein. Then molecular dynamics (MD) simulations in lipid bilayers are used to pinpoint likely lipid-protein interactions, which can be tested experimentally using HDX-MS. By bringing together the MD predictions with the conformational readouts from HDX-MS, we have uncovered key lipid-protein interactions implicated in stabilizing important functional conformations. This protocol can be applied to virtually any integral membrane protein amenable to classic biophysical studies and for which a near-atomic-resolution structure or homology model is available. This protocol takes ~4 d to complete, excluding the time for data analysis and MD simulations, which depends on the size of the protein under investigation.
中文翻译:

将氢-氘交换质谱与分子动力学模拟相结合,以探测膜蛋白中脂质调节的构象变化。
生物膜定义了细胞的边界,主要由磷脂和膜蛋白组成。越来越明显的是,膜蛋白与其周围脂质的直接相互作用在调节蛋白质构象和功能方面发挥着关键作用。然而,这些相互作用的确切性质和结构后果仍然难以在分子水平上追踪。在这里,我们提出了一个专门解决这一挑战的协议。首先,使用掺入受控脂质成分的纳米盘中的膜蛋白的氢-氘交换质谱(HDX-MS)来获得有关参与调节膜蛋白构象变化的脂质种类的信息。然后,利用脂质双层的分子动力学 (MD) 模拟来查明可能的脂质-蛋白质相互作用,并可以使用 HDX-MS 进行实验测试。通过将 MD 预测与 HDX-MS 的构象读数结合在一起,我们发现了与稳定重要功能构象有关的关键脂质-蛋白质相互作用。该协议几乎可以应用于任何适合经典生物物理学研究的整合膜蛋白,并且可以使用近原子分辨率的结构或同源模型。该协议需要大约 4 天才能完成,不包括数据分析和 MD 模拟的时间,这取决于所研究的蛋白质的大小。
更新日期:2019-10-12
中文翻译:

将氢-氘交换质谱与分子动力学模拟相结合,以探测膜蛋白中脂质调节的构象变化。
生物膜定义了细胞的边界,主要由磷脂和膜蛋白组成。越来越明显的是,膜蛋白与其周围脂质的直接相互作用在调节蛋白质构象和功能方面发挥着关键作用。然而,这些相互作用的确切性质和结构后果仍然难以在分子水平上追踪。在这里,我们提出了一个专门解决这一挑战的协议。首先,使用掺入受控脂质成分的纳米盘中的膜蛋白的氢-氘交换质谱(HDX-MS)来获得有关参与调节膜蛋白构象变化的脂质种类的信息。然后,利用脂质双层的分子动力学 (MD) 模拟来查明可能的脂质-蛋白质相互作用,并可以使用 HDX-MS 进行实验测试。通过将 MD 预测与 HDX-MS 的构象读数结合在一起,我们发现了与稳定重要功能构象有关的关键脂质-蛋白质相互作用。该协议几乎可以应用于任何适合经典生物物理学研究的整合膜蛋白,并且可以使用近原子分辨率的结构或同源模型。该协议需要大约 4 天才能完成,不包括数据分析和 MD 模拟的时间,这取决于所研究的蛋白质的大小。













































 京公网安备 11010802027423号
京公网安备 11010802027423号