Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Genotyping-by-sequencing based QTL mapping for rice grain yield under reproductive stage drought stress tolerance.
Scientific Reports ( IF 3.8 ) Pub Date : 2019-10-04 , DOI: 10.1038/s41598-019-50880-z Shailesh Yadav 1 , Nitika Sandhu 1, 2 , Vikas Kumar Singh 3 , Margaret Catolos 1 , Arvind Kumar 1, 4
Scientific Reports ( IF 3.8 ) Pub Date : 2019-10-04 , DOI: 10.1038/s41598-019-50880-z Shailesh Yadav 1 , Nitika Sandhu 1, 2 , Vikas Kumar Singh 3 , Margaret Catolos 1 , Arvind Kumar 1, 4
Affiliation
|
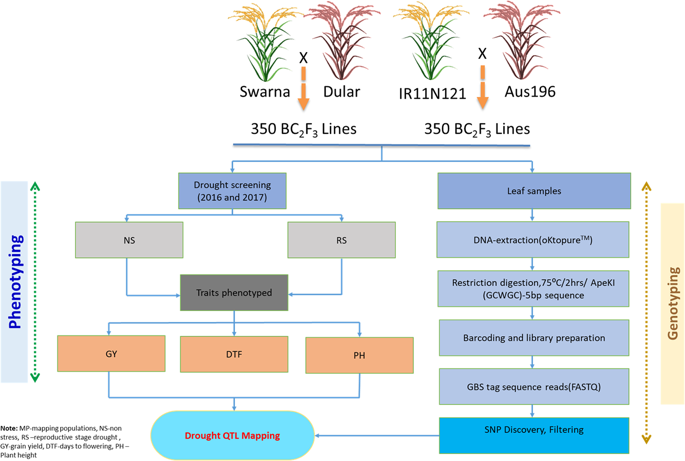
QTLs for rice grain yield under reproductive stage drought stress (qDTY) identified earlier with low density markers have shown linkage drag and need to be fine mapped before their utilization in breeding programs. In this study, genotyping-by-sequencing (GBS) based high-density linkage map of rice was developed using two BC1F3 mapping populations namely Swarna*2/Dular (3929 SNPs covering 1454.68 cM) and IR11N121*2/Aus196 (1191 SNPs covering 1399.68 cM) with average marker density of 0.37 cM to 1.18 cM respectively. In total, six qDTY QTLs including three consistent effect QTLs were identified in Swarna*2/Dular while eight qDTY QTLs including two consistent effect QTLs were identified in IR11N121*2/Aus 196 mapping population. Comparative analysis revealed four stable and novel QTLs (qDTY2.4, qDTY3.3, qDTY6.3, and qDTY11.2) which explained 8.62 to 14.92% PVE. However, one of the identified stable grain yield QTL qDTY1.1 in both the populations was located nearly at the same physical position of an earlier mapped major qDTY QTL. Further, the effect of the identified qDTY1.1 was validated in a subset of lines derived from five mapping populations confirming robustness of qDTY1.1 across various genetic backgrounds/seasons. The study successfully identified stable grain yield QTLs free from undesirable linkages of tall plant height/early maturity utilizing high density linkage maps.
中文翻译:

基于基因分型的QTL定位技术用于水稻生育期干旱胁迫耐受性的QTL定位。
早先用低密度标记确定的在生育期干旱胁迫下的水稻籽粒产量的QTL显示了连锁阻力,需要在作育计划之前加以精细定位。在这项研究中,使用两个BC1F3作图种群(即Swarna * 2 / Dular(3929个SNP,覆盖1454.68 cM)和IR11N121 * 2 / Aus196(1191个SNP),开发了基于基因分型(GBS)的水稻高密度连锁图谱。 1399.68 cM),平均标记密度分别为0.37 cM至1.18 cM。总共在Swarna * 2 / Dular中鉴定了6个qDTY QTL,包括3个一致效应QTL,而在IR11N121 * 2 / Aus 196制图群体中鉴定了8个qDTY QTL,包括2个一致效应QTL。对比分析显示了四个稳定且新颖的QTL(qDTY2.4,qDTY3.3,qDTY6.3和qDTY11.2),它们解释了8。PVE为62%至14.92%。但是,两个种群中已确定的稳定谷物产量QTL qDTY1.1之一几乎位于较早绘制的主要qDTY QTL的相同物理位置。此外,已鉴定的qDTY1.1的作用已在来自五个作图群体的一组品系中得到验证,从而确认了qDTY1.1在各种遗传背景/季节中的稳健性。该研究成功地利用高密度连锁图谱确定了无高株高/早熟不良连锁关系的稳定谷物产量QTL。在五个图谱种群衍生的品系子集中验证了1,从而确认了qDTY1.1在各种遗传背景/季节中的稳健性。该研究成功地利用高密度连锁图谱确定了无高株高/早熟不良连锁关系的稳定谷物产量QTL。在五个图谱种群衍生的品系子集中验证了1,从而确认了qDTY1.1在各种遗传背景/季节中的稳健性。该研究成功地利用高密度连锁图谱确定了无高株高/早熟不良连锁关系的稳定谷物产量QTL。
更新日期:2019-10-04
中文翻译:

基于基因分型的QTL定位技术用于水稻生育期干旱胁迫耐受性的QTL定位。
早先用低密度标记确定的在生育期干旱胁迫下的水稻籽粒产量的QTL显示了连锁阻力,需要在作育计划之前加以精细定位。在这项研究中,使用两个BC1F3作图种群(即Swarna * 2 / Dular(3929个SNP,覆盖1454.68 cM)和IR11N121 * 2 / Aus196(1191个SNP),开发了基于基因分型(GBS)的水稻高密度连锁图谱。 1399.68 cM),平均标记密度分别为0.37 cM至1.18 cM。总共在Swarna * 2 / Dular中鉴定了6个qDTY QTL,包括3个一致效应QTL,而在IR11N121 * 2 / Aus 196制图群体中鉴定了8个qDTY QTL,包括2个一致效应QTL。对比分析显示了四个稳定且新颖的QTL(qDTY2.4,qDTY3.3,qDTY6.3和qDTY11.2),它们解释了8。PVE为62%至14.92%。但是,两个种群中已确定的稳定谷物产量QTL qDTY1.1之一几乎位于较早绘制的主要qDTY QTL的相同物理位置。此外,已鉴定的qDTY1.1的作用已在来自五个作图群体的一组品系中得到验证,从而确认了qDTY1.1在各种遗传背景/季节中的稳健性。该研究成功地利用高密度连锁图谱确定了无高株高/早熟不良连锁关系的稳定谷物产量QTL。在五个图谱种群衍生的品系子集中验证了1,从而确认了qDTY1.1在各种遗传背景/季节中的稳健性。该研究成功地利用高密度连锁图谱确定了无高株高/早熟不良连锁关系的稳定谷物产量QTL。在五个图谱种群衍生的品系子集中验证了1,从而确认了qDTY1.1在各种遗传背景/季节中的稳健性。该研究成功地利用高密度连锁图谱确定了无高株高/早熟不良连锁关系的稳定谷物产量QTL。

































 京公网安备 11010802027423号
京公网安备 11010802027423号