当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Selection of 2'-Deoxy-2'-Fluoroarabino Nucleic Acid (FANA) Aptamers That Bind HIV-1 Integrase with Picomolar Affinity.
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2019-10-07 , DOI: 10.1021/acschembio.9b00237 Kevin M Rose 1 , Irani Alves Ferreira-Bravo 1 , Min Li 2 , Robert Craigie 2 , Mark A Ditzler 3 , Philipp Holliger 4 , Jeffrey J DeStefano 1, 5
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2019-10-07 , DOI: 10.1021/acschembio.9b00237 Kevin M Rose 1 , Irani Alves Ferreira-Bravo 1 , Min Li 2 , Robert Craigie 2 , Mark A Ditzler 3 , Philipp Holliger 4 , Jeffrey J DeStefano 1, 5
Affiliation
|
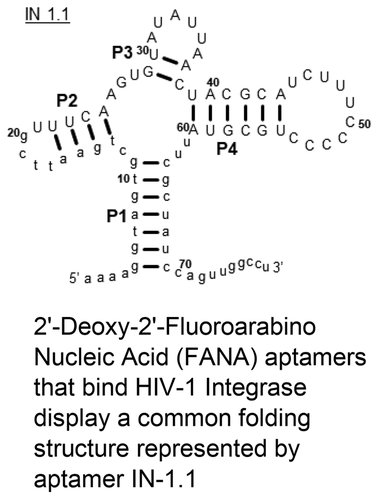
Systematic Evolution of Ligands by Exponential Enrichment (SELEX) is the iterative process by which nucleic acids that can bind with high affinity and specificity (termed aptamers) to specific protein targets are selected. Using a SELEX protocol adapted for Xeno-Nucleic Acid (XNA) as a suitable substrate for aptamer generation, 2'-fluoroarabinonucleic acid (FANA) was used to select several related aptamers to HIV-1 integrase (IN). IN bound FANA aptamers with equilibrium dissociation constants (KD,app) of ∼50-100 pM in a buffer with 200 mM NaCl and 6 mM MgCl2. Comparisons to published HIV-1 IN RNA and DNA aptamers as well as IN genomic binding partners indicated that FANA aptamers bound more than 2 orders of magnitude more tightly to IN. Using a combination of RNA folding algorithms and covariation analysis, all strong binding aptamers demonstrated a common four-way junction structure, despite significant sequence variation. IN aptamers were selected from the same starting library as FA1, a FANA aptamer that binds with pM affinity to HIV-1 Reverse Transcriptase (RT). It contains a 20-nucleotide 5' DNA sequence followed by 59 FANA nucleotides. IN-1.1 (one of the selected aptamers) potently inhibited IN activity and intasome formation in vitro. Replacing the FANA nucleotides of IN-1.1 with 2'-fluororibonucleic acid (F-RNA), which has the same chemical formula but with a ribose rather than arabinose sugar conformation, dramatically reduced binding, suggesting that FANA adopts unique structural conformations that promote binding to HIV-1 IN.
中文翻译:

结合HIV-1与皮摩尔亲和力的2'-脱氧-2'-氟阿拉伯糖核酸(FANA)适体的选择。
通过指数富集(SELEX)进行配体的系统进化是一个迭代过程,通过该过程可以选择能够以高亲和力和特异性结合到特定蛋白质靶标上的核酸。使用适合异种核酸(XNA)的SELEX方案作为适体产生的合适底物,使用2'-氟阿拉伯糖核酸(FANA)选择HIV-1整合酶(IN)的几种相关适体。在含有200 mM NaCl和6 mM MgCl2的缓冲液中,IN结合的FANA适体的平衡解离常数(KD,app)为〜50-100 pM。与已发表的HIV-1 IN RNA和DNA适体以及IN基因组结合伴侣的比较表明,FANA适体与IN的结合更紧密2个数量级。结合使用RNA折叠算法和协变分析,尽管有显着的序列变化,所有强结合适体均表现出共同的四向连接结构。IN适体选自与FA1相同的起始文库,FA1是与HIV-1逆转录酶(RT)以pM亲和力结合的FANA适体。它包含20个核苷酸的5'DNA序列,后跟59个FANA核苷酸。IN-1.1(选定的适体之一)在体外可有效抑制IN活性和基因体形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。IN适体选自与FA1相同的起始文库,FA1是与HIV-1逆转录酶(RT)以pM亲和力结合的FANA适体。它包含20个核苷酸的5'DNA序列,后跟59个FANA核苷酸。IN-1.1(选定的适体之一)在体外可有效抑制IN活性和基因体形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。IN适体选自与FA1相同的起始文库,FA1是与HIV-1逆转录酶(RT)以pM亲和力结合的FANA适体。它包含20个核苷酸的5'DNA序列,后跟59个FANA核苷酸。IN-1.1(选定的适体之一)在体外可有效抑制IN活性和基因体形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。1(选定的适体之一)在体外有效抑制IN活性和intasome形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。1(选定的适体之一)在体外有效抑制IN活性和intasome形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。
更新日期:2019-10-08
中文翻译:

结合HIV-1与皮摩尔亲和力的2'-脱氧-2'-氟阿拉伯糖核酸(FANA)适体的选择。
通过指数富集(SELEX)进行配体的系统进化是一个迭代过程,通过该过程可以选择能够以高亲和力和特异性结合到特定蛋白质靶标上的核酸。使用适合异种核酸(XNA)的SELEX方案作为适体产生的合适底物,使用2'-氟阿拉伯糖核酸(FANA)选择HIV-1整合酶(IN)的几种相关适体。在含有200 mM NaCl和6 mM MgCl2的缓冲液中,IN结合的FANA适体的平衡解离常数(KD,app)为〜50-100 pM。与已发表的HIV-1 IN RNA和DNA适体以及IN基因组结合伴侣的比较表明,FANA适体与IN的结合更紧密2个数量级。结合使用RNA折叠算法和协变分析,尽管有显着的序列变化,所有强结合适体均表现出共同的四向连接结构。IN适体选自与FA1相同的起始文库,FA1是与HIV-1逆转录酶(RT)以pM亲和力结合的FANA适体。它包含20个核苷酸的5'DNA序列,后跟59个FANA核苷酸。IN-1.1(选定的适体之一)在体外可有效抑制IN活性和基因体形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。IN适体选自与FA1相同的起始文库,FA1是与HIV-1逆转录酶(RT)以pM亲和力结合的FANA适体。它包含20个核苷酸的5'DNA序列,后跟59个FANA核苷酸。IN-1.1(选定的适体之一)在体外可有效抑制IN活性和基因体形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。IN适体选自与FA1相同的起始文库,FA1是与HIV-1逆转录酶(RT)以pM亲和力结合的FANA适体。它包含20个核苷酸的5'DNA序列,后跟59个FANA核苷酸。IN-1.1(选定的适体之一)在体外可有效抑制IN活性和基因体形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。1(选定的适体之一)在体外有效抑制IN活性和intasome形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。1(选定的适体之一)在体外有效抑制IN活性和intasome形成。用具有相同化学式但具有核糖而非阿拉伯糖构象的2'-氟核糖核酸(F-RNA)取代IN-1.1的FANA核苷酸,从而大大降低了结合,表明FANA采用了独特的结构构象来促进结合感染HIV-1 IN。






























 京公网安备 11010802027423号
京公网安备 11010802027423号