当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Validation of the Generalized Force Fields GAFF, CGenFF, OPLS-AA, and PRODRGFF by Testing Against Experimental Osmotic Coefficient Data for Small Drug-Like Molecules.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2019-10-08 , DOI: 10.1021/acs.jcim.9b00552 Shun Zhu 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2019-10-08 , DOI: 10.1021/acs.jcim.9b00552 Shun Zhu 1
Affiliation
|
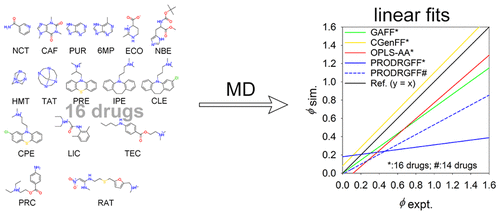
Accurate force field parameters for proteins and drugs are of fundamental importance for predicting protein-drug binding poses and binding free energies in structure-based drug design. Osmotic pressure simulations have recently gained popularity as a means to validate common force fields for biomolecules such as proteins; up to now, however, little such effort has been made to use these methods to validate common force fields for small drug-like molecules. In this work, osmotic coefficients of 16 drug-like molecules were calculated from osmotic pressure molecular dynamics simulations using four common force fields for drug-like molecules: GAFF, CGenFF, OPLS-AA, and PRODRGFF. While GAFF, CGenFF, and OPLS-AA produced osmotic coefficients that are in good agreement with the corresponding experimental osmotic coefficients, PRODRGFF, which attempts to use GROMOS parameters, often produced osmotic coefficients that are in poor agreement. All four force fields poorly reproduced the experimental osmotic coefficients of purine-derived molecules including purine, 6-methylpurine, and caffeine, suggesting common issues in describing interactions of this particular molecule type. The poor overall performance of PRODRGFF can be mainly attributed to the poor results with two macrocyclic molecules HMT and TAT, which can be significantly improved by reparameterizing the van der Waals (vdW) parameters of alkyl carbons and using lower solute concentration. In addition, the recently developed GAFF2 with revamped vdW parameters was found to produce osmotic coefficients that are in slightly better agreement with experiments than GAFF. Overall, the four common force fields for drug-like molecules tested in this study performed reasonably well at reproducing experimental osmotic coefficients of drug-like molecules, although failures of certain force fields on certain molecules suggest that further force field reparameterizations, especially for vdW parameters, might be required to improve their accuracy.
中文翻译:

通过针对类似小分子药物的实验渗透系数数据进行测试,验证了通用力场GAFF,CGenFF,OPLS-AA和PRODRGFF。
蛋白质和药物的准确力场参数对于预测基于结构的药物设计中的蛋白质-药物结合姿势和结合自由能至关重要。渗透压模拟作为一种验证诸如蛋白质之类的生物分子的共同力场的方法,最近已广受欢迎。然而,到目前为止,使用这些方法来验证类似药物的小分子的共同力场的努力还很少。在这项工作中,使用四种常见的药物样分子力场:GAFF,CGenFF,OPLS-AA和PRODRGFF,根据渗透压分子动力学模拟计算了16种药物样分子的渗透系数。虽然GAFF,CGenFF和OPLS-AA产生的渗透系数与相应的实验渗透系数PRODRGFF非常一致,它尝试使用GROMOS参数,经常产生渗透系数不一致的渗透系数。所有这四个力场都无法很好地再现嘌呤,6-甲基嘌呤和咖啡因等嘌呤衍生分子的实验渗透系数,这在描述这种特定分子类型的相互作用时提出了常见问题。PRODRGFF总体性能差的主要原因是两个大环分子HMT和TAT的结果差,这可以通过重新参数化烷基碳的范德华(vdW)参数并使用较低的溶质浓度来显着改善。此外,发现最近开发的带有修改过的vdW参数的GAFF2产生的渗透系数与实验相比要好于GAFF。全面的,
更新日期:2019-10-10
中文翻译:

通过针对类似小分子药物的实验渗透系数数据进行测试,验证了通用力场GAFF,CGenFF,OPLS-AA和PRODRGFF。
蛋白质和药物的准确力场参数对于预测基于结构的药物设计中的蛋白质-药物结合姿势和结合自由能至关重要。渗透压模拟作为一种验证诸如蛋白质之类的生物分子的共同力场的方法,最近已广受欢迎。然而,到目前为止,使用这些方法来验证类似药物的小分子的共同力场的努力还很少。在这项工作中,使用四种常见的药物样分子力场:GAFF,CGenFF,OPLS-AA和PRODRGFF,根据渗透压分子动力学模拟计算了16种药物样分子的渗透系数。虽然GAFF,CGenFF和OPLS-AA产生的渗透系数与相应的实验渗透系数PRODRGFF非常一致,它尝试使用GROMOS参数,经常产生渗透系数不一致的渗透系数。所有这四个力场都无法很好地再现嘌呤,6-甲基嘌呤和咖啡因等嘌呤衍生分子的实验渗透系数,这在描述这种特定分子类型的相互作用时提出了常见问题。PRODRGFF总体性能差的主要原因是两个大环分子HMT和TAT的结果差,这可以通过重新参数化烷基碳的范德华(vdW)参数并使用较低的溶质浓度来显着改善。此外,发现最近开发的带有修改过的vdW参数的GAFF2产生的渗透系数与实验相比要好于GAFF。全面的,

































 京公网安备 11010802027423号
京公网安备 11010802027423号