Nature Biotechnology ( IF 33.1 ) Pub Date : 2019-09-09 , DOI: 10.1038/s41587-019-0240-x Leon Anavy , Inbal Vaknin , Orna Atar , Roee Amit , Zohar Yakhini
|
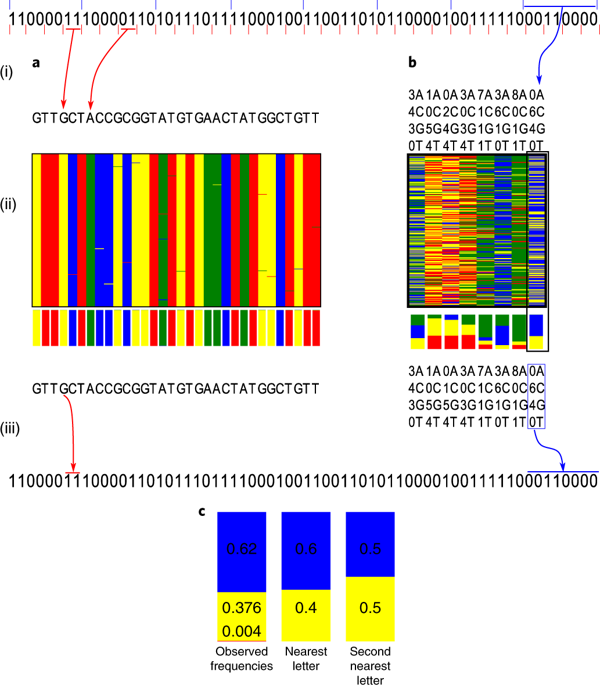
The density and long-term stability of DNA make it an appealing storage medium, particularly for long-term data archiving. Existing DNA storage technologies involve the synthesis and sequencing of multiple nominally identical molecules in parallel, resulting in information redundancy. We report the development of encoding and decoding methods that exploit this redundancy using composite DNA letters. A composite DNA letter is a representation of a position in a sequence that consists of a mixture of all four DNA nucleotides in a predetermined ratio. Our methods encode data using fewer synthesis cycles. We encode 6.4 MB into composite DNA, with distinguishable composition medians, using 20% fewer synthesis cycles per unit of data, as compared to previous reports. We also simulate encoding with larger composite alphabets, with distinguishable composition deciles, to show that 75% fewer synthesis cycles are potentially sufficient. We describe applicable error-correcting codes and inference methods, and investigate error patterns in the context of composite DNA letters.
中文翻译:

使用复合DNA字母以更少的合成周期将数据存储在DNA中
DNA的密度和长期稳定性使其成为一种有吸引力的存储介质,特别是对于长期数据归档而言。现有的DNA存储技术涉及多个名义上相同的分子的并行合成和测序,从而导致信息冗余。我们报告了利用复合DNA字母利用这种冗余的编码和解码方法的发展。复合DNA字母表示序列中某个位置的位置,该序列由所有四个DNA核苷酸以预定比例的混合物组成。我们的方法使用更少的合成周期对数据进行编码。与以前的报告相比,我们将6.4 MB编码为复合DNA,具有可区分的组成中位数,每单位数据使用的合成周期减少了20%。我们还使用较大的复合字母来模拟编码,具有可分辨的组成十分之一,表明减少75%的合成循环可能就足够了。我们描述了适用的纠错码和推断方法,并研究了复合DNA字母中的错误模式。













































 京公网安备 11010802027423号
京公网安备 11010802027423号