Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape
Nature ( IF 50.5 ) Pub Date : 2019-09-04 , DOI: 10.1038/s41586-019-1534-3 Daniel N Weinberg 1 , Simon Papillon-Cavanagh 2 , Haifen Chen 2 , Yuan Yue 3 , Xiao Chen 4, 5 , Kartik N Rajagopalan 6 , Cynthia Horth 2 , John T McGuire 4, 5 , Xinjing Xu 4, 5 , Hamid Nikbakht 2 , Agata E Lemiesz 1 , Dylan M Marchione 7, 8 , Matthew R Marunde 9 , Matthew J Meiners 9 , Marcus A Cheek 9 , Michael-Christopher Keogh 9 , Eric Bareke 2 , Anissa Djedid 2 , Ashot S Harutyunyan 2 , Nada Jabado 2, 10, 11 , Benjamin A Garcia 7, 8 , Haitao Li 3 , C David Allis 1 , Jacek Majewski 2 , Chao Lu 4, 5
Nature ( IF 50.5 ) Pub Date : 2019-09-04 , DOI: 10.1038/s41586-019-1534-3 Daniel N Weinberg 1 , Simon Papillon-Cavanagh 2 , Haifen Chen 2 , Yuan Yue 3 , Xiao Chen 4, 5 , Kartik N Rajagopalan 6 , Cynthia Horth 2 , John T McGuire 4, 5 , Xinjing Xu 4, 5 , Hamid Nikbakht 2 , Agata E Lemiesz 1 , Dylan M Marchione 7, 8 , Matthew R Marunde 9 , Matthew J Meiners 9 , Marcus A Cheek 9 , Michael-Christopher Keogh 9 , Eric Bareke 2 , Anissa Djedid 2 , Ashot S Harutyunyan 2 , Nada Jabado 2, 10, 11 , Benjamin A Garcia 7, 8 , Haitao Li 3 , C David Allis 1 , Jacek Majewski 2 , Chao Lu 4, 5
Affiliation
|
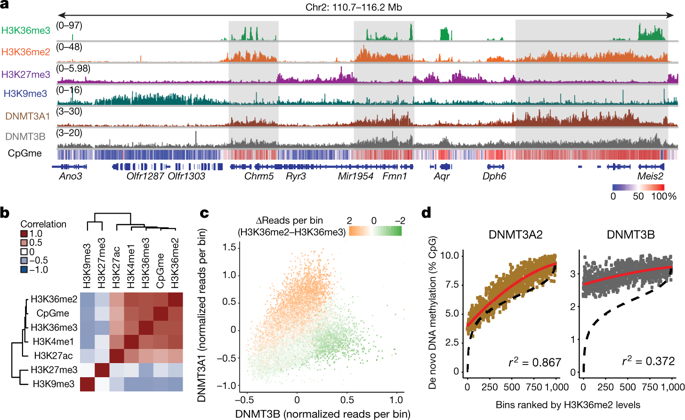
Enzymes that catalyse CpG methylation in DNA, including the DNA methyltransferases 1 (DNMT1), 3A (DNMT3A) and 3B (DNMT3B), are indispensable for mammalian tissue development and homeostasis1–4. They are also implicated in human developmental disorders and cancers5–8, supporting the critical role of DNA methylation in the specification and maintenance of cell fate. Previous studies have suggested that post-translational modifications of histones are involved in specifying patterns of DNA methyltransferase localization and DNA methylation at promoters and actively transcribed gene bodies9–11. However, the mechanisms that control the establishment and maintenance of intergenic DNA methylation remain poorly understood. Tatton–Brown–Rahman syndrome (TBRS) is a childhood overgrowth disorder that is defined by germline mutations in DNMT3A. TBRS shares clinical features with Sotos syndrome (which is caused by haploinsufficiency of NSD1, a histone methyltransferase that catalyses the dimethylation of histone H3 at K36 (H3K36me2)8,12,13), which suggests that there is a mechanistic link between these two diseases. Here we report that NSD1-mediated H3K36me2 is required for the recruitment of DNMT3A and maintenance of DNA methylation at intergenic regions. Genome-wide analysis shows that the binding and activity of DNMT3A colocalize with H3K36me2 at non-coding regions of euchromatin. Genetic ablation of Nsd1 and its paralogue Nsd2 in mouse cells results in a redistribution of DNMT3A to H3K36me3-modified gene bodies and a reduction in the methylation of intergenic DNA. Blood samples from patients with Sotos syndrome and NSD1-mutant tumours also exhibit hypomethylation of intergenic DNA. The PWWP domain of DNMT3A shows dual recognition of H3K36me2 and H3K36me3 in vitro, with a higher binding affinity towards H3K36me2 that is abrogated by TBRS-derived missense mutations. Together, our study reveals a trans-chromatin regulatory pathway that connects aberrant intergenic CpG methylation to human neoplastic and developmental overgrowth.H3K36me2 targets DNMT3A to intergenic regions and this process, together with H3K36me3-mediated recruitment of DNMT3B, has a key role in establishing and maintaining genomic DNA methylation landscapes.
中文翻译:

组蛋白标记 H3K36me2 招募 DNMT3A 并塑造基因间 DNA 甲基化景观
催化 DNA 中 CpG 甲基化的酶,包括 DNA 甲基转移酶 1 (DNMT1)、3A (DNMT3A) 和 3B (DNMT3B),对于哺乳动物组织发育和体内平衡是必不可少的1-4。它们还涉及人类发育障碍和癌症 5-8,支持 DNA 甲基化在细胞命运的规范和维持中的关键作用。以前的研究表明,组蛋白的翻译后修饰参与指定启动子和主动转录基因体的 DNA 甲基转移酶定位和 DNA 甲基化模式9-11。然而,控制基因间 DNA 甲基化建立和维持的机制仍然知之甚少。Tatton-Brown-Rahman 综合征 (TBRS) 是一种儿童过度生长疾病,由 DNMT3A 中的种系突变定义。TBRS 与 Sotos 综合征(由 NSD1 的单倍剂量不足引起,NSD1 是一种组蛋白甲基转移酶,催化组蛋白 H3 在 K36 (H3K36me2)8,12,13 处二甲基化)的临床特征,这表明这两种疾病之间存在机制联系. 在这里我们报告 NSD1 介导的 H3K36me2 是招募 DNMT3A 和维持基因间区域 DNA 甲基化所必需的。全基因组分析表明,DNMT3A 的结合和活性与 H3K36me2 共定位于常染色质的非编码区。小鼠细胞中 Nsd1 及其旁系同源物 Nsd2 的遗传消融导致 DNMT3A 重新分布到 H3K36me3 修饰的基因体,并减少基因间 DNA 的甲基化。来自 Sotos 综合征和 NSD1 突变肿瘤患者的血液样本也表现出基因间 DNA 的低甲基化。DNMT3A 的 PWWP 域在体外显示出对 H3K36me2 和 H3K36me3 的双重识别,对 H3K36me2 具有更高的结合亲和力,这被 TBRS 衍生的错义突变消除。总之,我们的研究揭示了将异常基因间 CpG 甲基化与人类肿瘤和发育过度生长联系起来的转染色质调控途径。 H3K36me2 将 DNMT3A 靶向基因间区域,这个过程与 H3K36me3 介导的 DNMT3B 募集一起,在建立和维持基因组 DNA 甲基化景观。
更新日期:2019-09-04
中文翻译:

组蛋白标记 H3K36me2 招募 DNMT3A 并塑造基因间 DNA 甲基化景观
催化 DNA 中 CpG 甲基化的酶,包括 DNA 甲基转移酶 1 (DNMT1)、3A (DNMT3A) 和 3B (DNMT3B),对于哺乳动物组织发育和体内平衡是必不可少的1-4。它们还涉及人类发育障碍和癌症 5-8,支持 DNA 甲基化在细胞命运的规范和维持中的关键作用。以前的研究表明,组蛋白的翻译后修饰参与指定启动子和主动转录基因体的 DNA 甲基转移酶定位和 DNA 甲基化模式9-11。然而,控制基因间 DNA 甲基化建立和维持的机制仍然知之甚少。Tatton-Brown-Rahman 综合征 (TBRS) 是一种儿童过度生长疾病,由 DNMT3A 中的种系突变定义。TBRS 与 Sotos 综合征(由 NSD1 的单倍剂量不足引起,NSD1 是一种组蛋白甲基转移酶,催化组蛋白 H3 在 K36 (H3K36me2)8,12,13 处二甲基化)的临床特征,这表明这两种疾病之间存在机制联系. 在这里我们报告 NSD1 介导的 H3K36me2 是招募 DNMT3A 和维持基因间区域 DNA 甲基化所必需的。全基因组分析表明,DNMT3A 的结合和活性与 H3K36me2 共定位于常染色质的非编码区。小鼠细胞中 Nsd1 及其旁系同源物 Nsd2 的遗传消融导致 DNMT3A 重新分布到 H3K36me3 修饰的基因体,并减少基因间 DNA 的甲基化。来自 Sotos 综合征和 NSD1 突变肿瘤患者的血液样本也表现出基因间 DNA 的低甲基化。DNMT3A 的 PWWP 域在体外显示出对 H3K36me2 和 H3K36me3 的双重识别,对 H3K36me2 具有更高的结合亲和力,这被 TBRS 衍生的错义突变消除。总之,我们的研究揭示了将异常基因间 CpG 甲基化与人类肿瘤和发育过度生长联系起来的转染色质调控途径。 H3K36me2 将 DNMT3A 靶向基因间区域,这个过程与 H3K36me3 介导的 DNMT3B 募集一起,在建立和维持基因组 DNA 甲基化景观。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号