当前位置:
X-MOL 学术
›
Nat. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Assessment of network module identification across complex diseases.
Nature Methods ( IF 36.1 ) Pub Date : 2019-08-30 , DOI: 10.1038/s41592-019-0509-5 Sarvenaz Choobdar 1, 2 , Mehmet E Ahsen 3 , Jake Crawford 4 , Mattia Tomasoni 1, 2 , Tao Fang 5 , David Lamparter 1, 2, 6 , Junyuan Lin 7 , Benjamin Hescott 8 , Xiaozhe Hu 7 , Johnathan Mercer 9, 10 , Ted Natoli 11 , Rajiv Narayan 11 , , Aravind Subramanian 11 , Jitao D Zhang 5 , Gustavo Stolovitzky 3, 12 , Zoltán Kutalik 2, 13 , Kasper Lage 9, 10, 14 , Donna K Slonim 4, 15 , Julio Saez-Rodriguez 16, 17 , Lenore J Cowen 4, 7 , Sven Bergmann 1, 2, 18 , Daniel Marbach 1, 2, 5
Nature Methods ( IF 36.1 ) Pub Date : 2019-08-30 , DOI: 10.1038/s41592-019-0509-5 Sarvenaz Choobdar 1, 2 , Mehmet E Ahsen 3 , Jake Crawford 4 , Mattia Tomasoni 1, 2 , Tao Fang 5 , David Lamparter 1, 2, 6 , Junyuan Lin 7 , Benjamin Hescott 8 , Xiaozhe Hu 7 , Johnathan Mercer 9, 10 , Ted Natoli 11 , Rajiv Narayan 11 , , Aravind Subramanian 11 , Jitao D Zhang 5 , Gustavo Stolovitzky 3, 12 , Zoltán Kutalik 2, 13 , Kasper Lage 9, 10, 14 , Donna K Slonim 4, 15 , Julio Saez-Rodriguez 16, 17 , Lenore J Cowen 4, 7 , Sven Bergmann 1, 2, 18 , Daniel Marbach 1, 2, 5
Affiliation
|
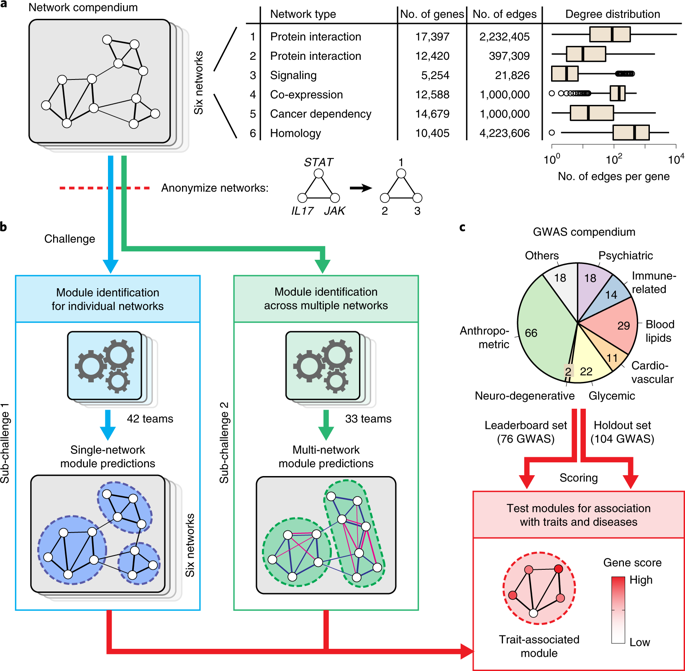
Many bioinformatics methods have been proposed for reducing the complexity of large gene or protein networks into relevant subnetworks or modules. Yet, how such methods compare to each other in terms of their ability to identify disease-relevant modules in different types of network remains poorly understood. We launched the 'Disease Module Identification DREAM Challenge', an open competition to comprehensively assess module identification methods across diverse protein-protein interaction, signaling, gene co-expression, homology and cancer-gene networks. Predicted network modules were tested for association with complex traits and diseases using a unique collection of 180 genome-wide association studies. Our robust assessment of 75 module identification methods reveals top-performing algorithms, which recover complementary trait-associated modules. We find that most of these modules correspond to core disease-relevant pathways, which often comprise therapeutic targets. This community challenge establishes biologically interpretable benchmarks, tools and guidelines for molecular network analysis to study human disease biology.
中文翻译:

跨复杂疾病的网络模块识别评估。
已经提出了许多生物信息学方法来将大基因或蛋白质网络的复杂性降低到相关子网络或模块中。然而,这些方法在识别不同类型网络中疾病相关模块的能力方面如何相互比较仍然知之甚少。我们发起了“疾病模块识别梦想挑战”,这是一项公开竞赛,旨在全面评估跨不同蛋白质-蛋白质相互作用、信号传导、基因共表达、同源性和癌症基因网络的模块识别方法。使用 180 个全基因组关联研究的独特集合来测试预测的网络模块与复杂性状和疾病的关联。我们对 75 种模块识别方法的稳健评估揭示了性能最佳的算法,这些算法可以恢复互补的特征相关模块。我们发现这些模块中的大多数对应于核心疾病相关途径,通常包含治疗靶点。这一社区挑战为分子网络分析建立了生物学上可解释的基准、工具和指南,以研究人类疾病生物学。
更新日期:2019-08-30
中文翻译:

跨复杂疾病的网络模块识别评估。
已经提出了许多生物信息学方法来将大基因或蛋白质网络的复杂性降低到相关子网络或模块中。然而,这些方法在识别不同类型网络中疾病相关模块的能力方面如何相互比较仍然知之甚少。我们发起了“疾病模块识别梦想挑战”,这是一项公开竞赛,旨在全面评估跨不同蛋白质-蛋白质相互作用、信号传导、基因共表达、同源性和癌症基因网络的模块识别方法。使用 180 个全基因组关联研究的独特集合来测试预测的网络模块与复杂性状和疾病的关联。我们对 75 种模块识别方法的稳健评估揭示了性能最佳的算法,这些算法可以恢复互补的特征相关模块。我们发现这些模块中的大多数对应于核心疾病相关途径,通常包含治疗靶点。这一社区挑战为分子网络分析建立了生物学上可解释的基准、工具和指南,以研究人类疾病生物学。































 京公网安备 11010802027423号
京公网安备 11010802027423号