Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules.
Molecular Cell ( IF 14.5 ) Pub Date : 2019-08-22 , DOI: 10.1016/j.molcel.2019.07.030
Alejandro Padrón 1 , Shintaro Iwasaki 2 , Nicholas T Ingolia 1
Molecular Cell ( IF 14.5 ) Pub Date : 2019-08-22 , DOI: 10.1016/j.molcel.2019.07.030
Alejandro Padrón 1 , Shintaro Iwasaki 2 , Nicholas T Ingolia 1
Affiliation
|
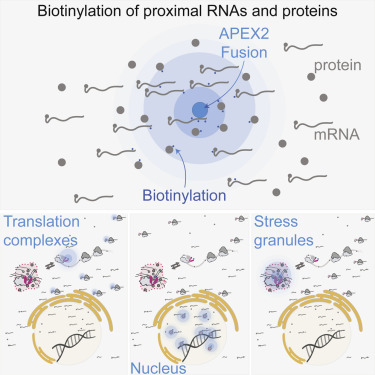
Diverse ribonucleoprotein complexes control mRNA processing, translation, and decay. Transcripts in these complexes localize to specific regions of the cell and can condense into non-membrane-bound structures such as stress granules. It has proven challenging to map the RNA composition of these large and dynamic structures, however. We therefore developed an RNA proximity labeling technique, APEX-seq, which uses the ascorbate peroxidase APEX2 to probe the spatial organization of the transcriptome. We show that APEX-seq can resolve the localization of RNAs within the cell and determine their enrichment or depletion near key RNA-binding proteins. Matching the spatial transcriptome, as revealed by APEX-seq, with the spatial proteome determined by APEX-mass spectrometry (APEX-MS), obtained precisely in parallel, provides new insights into the organization of translation initiation complexes on active mRNAs and unanticipated complexity in stress granule composition. Our novel technique allows a powerful and general approach to explore the spatial environment of macromolecules.
中文翻译:

由APEX-Seq进行的近端RNA标记揭示了翻译起始复合物和抑制性RNA颗粒的组织。
不同的核糖核蛋白复合物控制mRNA的加工,翻译和衰变。这些复合物中的转录物位于细胞的特定区域,并且可以凝结成非膜结合的结构,例如应力颗粒。然而,事实证明,绘制这些大而动态的结构的RNA组成具有挑战性。因此,我们开发了一种RNA邻近标记技术APEX-seq,它使用抗坏血酸过氧化物酶APEX2来探测转录组的空间组织。我们表明,APEX-seq可以解决细胞内RNA的定位,并确定其在关键RNA结合蛋白附近的富集或耗竭。将APEX-seq揭示的空间转录组与APEX-质谱(APEX-MS)确定的空间蛋白质组进行精确匹配,提供了有关活性mRNA上翻译起始复合物组织的新见解,以及应激颗粒组成中意料之外的复杂性。我们的新技术提供了一种强大而通用的方法来探索大分子的空间环境。
更新日期:2019-08-23
中文翻译:

由APEX-Seq进行的近端RNA标记揭示了翻译起始复合物和抑制性RNA颗粒的组织。
不同的核糖核蛋白复合物控制mRNA的加工,翻译和衰变。这些复合物中的转录物位于细胞的特定区域,并且可以凝结成非膜结合的结构,例如应力颗粒。然而,事实证明,绘制这些大而动态的结构的RNA组成具有挑战性。因此,我们开发了一种RNA邻近标记技术APEX-seq,它使用抗坏血酸过氧化物酶APEX2来探测转录组的空间组织。我们表明,APEX-seq可以解决细胞内RNA的定位,并确定其在关键RNA结合蛋白附近的富集或耗竭。将APEX-seq揭示的空间转录组与APEX-质谱(APEX-MS)确定的空间蛋白质组进行精确匹配,提供了有关活性mRNA上翻译起始复合物组织的新见解,以及应激颗粒组成中意料之外的复杂性。我们的新技术提供了一种强大而通用的方法来探索大分子的空间环境。

































 京公网安备 11010802027423号
京公网安备 11010802027423号