Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Reconstituting the transcriptome and DNA methylome landscapes of human implantation
Nature ( IF 50.5 ) Pub Date : 2019-08-21 , DOI: 10.1038/s41586-019-1500-0 Fan Zhou 1, 2 , Rui Wang 1, 2 , Peng Yuan 1, 3 , Yixin Ren 1, 3 , Yunuo Mao 1, 2 , Rong Li 1, 3 , Ying Lian 1, 3 , Junsheng Li 1, 3 , Lu Wen 1, 2 , Liying Yan 1, 2, 3, 4 , Jie Qiao 1, 2, 3, 4, 5, 6 , Fuchou Tang 1, 2, 3, 5, 6
Nature ( IF 50.5 ) Pub Date : 2019-08-21 , DOI: 10.1038/s41586-019-1500-0 Fan Zhou 1, 2 , Rui Wang 1, 2 , Peng Yuan 1, 3 , Yixin Ren 1, 3 , Yunuo Mao 1, 2 , Rong Li 1, 3 , Ying Lian 1, 3 , Junsheng Li 1, 3 , Lu Wen 1, 2 , Liying Yan 1, 2, 3, 4 , Jie Qiao 1, 2, 3, 4, 5, 6 , Fuchou Tang 1, 2, 3, 5, 6
Affiliation
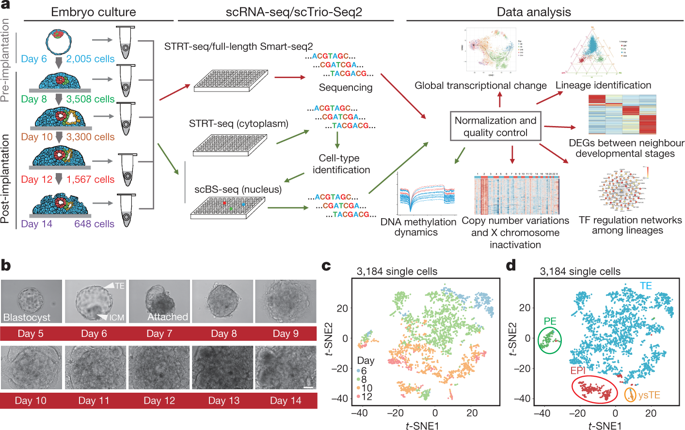
|
Implantation is a milestone event during mammalian embryogenesis. Implantation failure is a considerable cause of early pregnancy loss in humans1. Owing to the difficulty of obtaining human embryos early after implantation in vivo, it remains unclear how the gene regulatory network and epigenetic mechanisms control the implantation process. Here, by combining an in vitro culture system for the development human embryos after implantation and single-cell multi-omics sequencing technologies, more than 8,000 individual cells from 65 human peri-implantation embryos were systematically analysed. Unsupervised dimensionality reduction and clustering algorithms of the transcriptome data show stepwise implantation routes for the epiblast, primitive endoderm and trophectoderm lineages, suggesting robust preparation for the proper establishment of a mother-to-offspring connection during implantation. Female embryos showed initiation of random X chromosome inactivation based on analysis of parental allele-specific expression of X-chromosome-linked genes during implantation. Notably, using single-cell triple omics sequencing analysis, the re-methylation of the genome in cells from the primitive endoderm lineage was shown to be much slower than in cells of both epiblast and trophectoderm lineages during the implantation process, which indicates that there are distinct re-establishment features in the DNA methylome of the epiblast and primitive endoderm—even though both lineages are derived from the inner cell mass. Collectively, our work provides insights into the complex molecular mechanisms that regulate the implantation of human embryos, and helps to advance future efforts to understanding early embryonic development and reproductive medicine.Transcriptomics and DNA methylomics are used to study the implantation process of human embryos at single-cell resolution.
中文翻译:

重建人类植入的转录组和 DNA 甲基化组景观
植入是哺乳动物胚胎发生过程中的里程碑事件。着床失败是人类早期流产的一个重要原因1。由于在体内植入后早期难以获得人类胚胎,目前尚不清楚基因调控网络和表观遗传机制如何控制植入过程。在这里,通过结合体外发育人类胚胎植入后培养系统和单细胞多组学测序技术,系统地分析了来自 65 个人类着床周围胚胎的 8,000 多个单个细胞。转录组数据的无监督降维和聚类算法显示了外胚层、原始内胚层和滋养外胚层谱系的逐步植入途径,建议为在植入过程中正确建立母与子之间的联系做强有力的准备。根据对着床期间 X 染色体连锁基因的亲本等位基因特异性表达的分析,女性胚胎显示出随机 X 染色体失活的开始。值得注意的是,使用单细胞三重组学测序分析,原始内胚层谱系细胞中基因组的再甲基化显示出在植入过程中比外胚层和滋养外胚层谱系的细胞慢得多,这表明存在外胚层和原始内胚层的 DNA 甲基化组具有明显的重建特征——即使这两个谱系都来自内细胞团。总的来说,
更新日期:2019-08-21
中文翻译:

重建人类植入的转录组和 DNA 甲基化组景观
植入是哺乳动物胚胎发生过程中的里程碑事件。着床失败是人类早期流产的一个重要原因1。由于在体内植入后早期难以获得人类胚胎,目前尚不清楚基因调控网络和表观遗传机制如何控制植入过程。在这里,通过结合体外发育人类胚胎植入后培养系统和单细胞多组学测序技术,系统地分析了来自 65 个人类着床周围胚胎的 8,000 多个单个细胞。转录组数据的无监督降维和聚类算法显示了外胚层、原始内胚层和滋养外胚层谱系的逐步植入途径,建议为在植入过程中正确建立母与子之间的联系做强有力的准备。根据对着床期间 X 染色体连锁基因的亲本等位基因特异性表达的分析,女性胚胎显示出随机 X 染色体失活的开始。值得注意的是,使用单细胞三重组学测序分析,原始内胚层谱系细胞中基因组的再甲基化显示出在植入过程中比外胚层和滋养外胚层谱系的细胞慢得多,这表明存在外胚层和原始内胚层的 DNA 甲基化组具有明显的重建特征——即使这两个谱系都来自内细胞团。总的来说,




















































 京公网安备 11010802027423号
京公网安备 11010802027423号