Scientific Reports ( IF 3.8 ) Pub Date : 2019-07-17 , DOI: 10.1038/s41598-019-45938-x Daniel K Putnam 1 , Xiaotu Ma 1 , Stephen V Rice 1 , Yu Liu 1 , Scott Newman 1 , Jinghui Zhang 1 , Xiang Chen 1
|
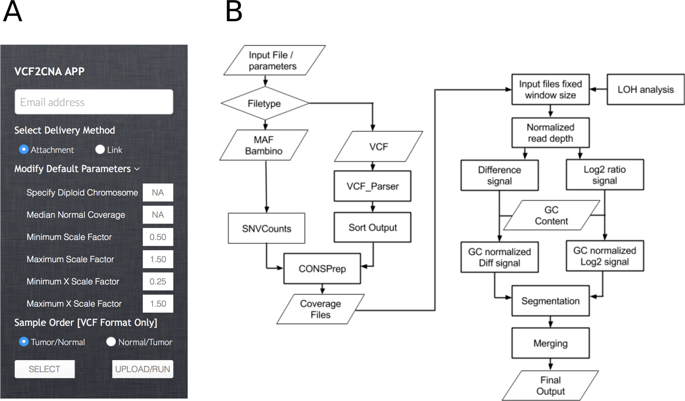
VCF2CNA is a tool (Linux commandline or web-interface) for copy-number alteration (CNA) analysis and tumor purity estimation of paired tumor-normal VCF variant file formats. It operates on whole genome and whole exome datasets. To benchmark its performance, we applied it to 46 adult glioblastoma and 146 pediatric neuroblastoma samples sequenced by Illumina and Complete Genomics (CGI) platforms respectively. VCF2CNA was highly consistent with a state-of-the-art algorithm using raw sequencing data (mean F1-score = 0.994) in high-quality whole genome glioblastoma samples and was robust to uneven coverage introduced by library artifacts. In the whole genome neuroblastoma set, VCF2CNA identified MYCN high-level amplifications in 31 of 32 clinically validated samples compared to 15 found by CGI’s HMM-based CNA model. Moreover, VCF2CNA achieved highly consistent CNA profiles between WGS and WXS platforms (mean F1 score 0.97 on a set of 15 rhabdomyosarcoma samples). In addition, VCF2CNA provides accurate tumor purity estimates for samples with sufficient CNAs. These results suggest that VCF2CNA is an accurate, efficient and platform-independent tool for CNA and tumor purity analyses without accessing raw sequence data.
中文翻译:

VCF2CNA:一种用于有效检测VCF基因型数据中拷贝数变化和肿瘤纯度的工具。
VCF2CNA是一个工具(Linux命令行或Web界面),用于成对的肿瘤正常VCF变异文件格式的拷贝数变更(CNA)分析和肿瘤纯度估计。它对整个基因组和整个外显子组数据集起作用。为了评估其性能,我们将其应用于分别通过Illumina和Complete Genomics(CGI)平台测序的46个成年胶质母细胞瘤和146个小儿神经母细胞瘤样品。VCF2CNA与在高质量全基因组胶质母细胞瘤样品中使用原始测序数据(均值F1分数= 0.994)的最新算法高度一致,并且对于库文物引入的不均匀覆盖具有鲁棒性。在整个基因组神经母细胞瘤中,VCF2CNA在32个经过临床验证的样本中有31个鉴定出MYCN高水平扩增,而CGI基于HMM的CNA模型发现了15个。而且,VCF2CNA在WGS和WXS平台之间实现了高度一致的CNA配置文件(一组15个横纹肌肉瘤样本的平均F1评分为0.97)。此外,VCF2CNA可为具有足够CNA的样品提供准确的肿瘤纯度估计。这些结果表明,VCF2CNA是用于CNA和肿瘤纯度分析的准确,高效且独立于平台的工具,而无需访问原始序列数据。






























 京公网安备 11010802027423号
京公网安备 11010802027423号