Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mapping the Mouse Cell Atlas by Microwell-Seq.
Cell ( IF 45.5 ) Pub Date : 2018-Feb-22 , DOI: 10.1016/j.cell.2018.02.001 Xiaoping Han , Renying Wang , Yincong Zhou , Lijiang Fei , Huiyu Sun , Shujing Lai , Assieh Saadatpour , Ziming Zhou , Haide Chen , Fang Ye , Daosheng Huang , Yang Xu , Wentao Huang , Mengmeng Jiang , Xinyi Jiang , Jie Mao , Yao Chen , Chenyu Lu , Jin Xie , Qun Fang , Yibin Wang , Rui Yue , Tiefeng Li , He Huang , Stuart H. Orkin , Guo-Cheng Yuan , Ming Chen , Guoji Guo
Cell ( IF 45.5 ) Pub Date : 2018-Feb-22 , DOI: 10.1016/j.cell.2018.02.001 Xiaoping Han , Renying Wang , Yincong Zhou , Lijiang Fei , Huiyu Sun , Shujing Lai , Assieh Saadatpour , Ziming Zhou , Haide Chen , Fang Ye , Daosheng Huang , Yang Xu , Wentao Huang , Mengmeng Jiang , Xinyi Jiang , Jie Mao , Yao Chen , Chenyu Lu , Jin Xie , Qun Fang , Yibin Wang , Rui Yue , Tiefeng Li , He Huang , Stuart H. Orkin , Guo-Cheng Yuan , Ming Chen , Guoji Guo
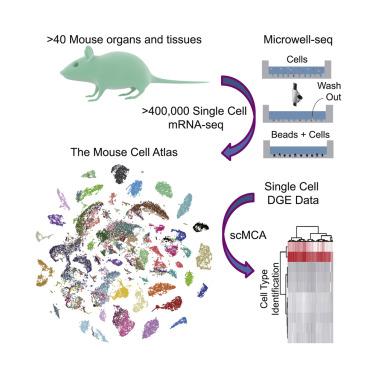
|
Single-cell RNA sequencing (scRNA-seq) technologies are poised to reshape the current cell-type classification system. However, a transcriptome-based single-cell atlas has not been achieved for complex mammalian systems. Here, we developed Microwell-seq, a high-throughput and low-cost scRNA-seq platform using simple, inexpensive devices. Using Microwell-seq, we analyzed more than 400,000 single cells covering all of the major mouse organs and constructed a basic scheme for a mouse cell atlas (MCA). We reveal a single-cell hierarchy for many tissues that have not been well characterized previously. We built a web-based "single-cell MCA analysis" pipeline that accurately defines cell types based on single-cell digital expression. Our study demonstrates the wide applicability of the Microwell-seq technology and MCA resource.
中文翻译:

通过Microwell-Seq绘制小鼠细胞图谱。
单细胞RNA测序(scRNA-seq)技术已准备好重塑当前的细胞类型分类系统。但是,对于复杂的哺乳动物系统,尚未实现基于转录组的单细胞图谱。在这里,我们使用简单,便宜的设备开发了Microwell-seq,一种高通量,低成本的scRNA-seq平台。使用Microwell-seq,我们分析了覆盖所有主要小鼠器官的400,000多个单细胞,并构建了小鼠细胞图谱(MCA)的基本方案。我们揭示了以前没有很好表征的许多组织的单细胞层次结构。我们建立了一个基于Web的“单细胞MCA分析”管道,该管道可根据单细胞数字表达准确定义细胞类型。我们的研究证明了微孔测序技术和MCA资源的广泛适用性。
更新日期:2018-02-22
中文翻译:

通过Microwell-Seq绘制小鼠细胞图谱。
单细胞RNA测序(scRNA-seq)技术已准备好重塑当前的细胞类型分类系统。但是,对于复杂的哺乳动物系统,尚未实现基于转录组的单细胞图谱。在这里,我们使用简单,便宜的设备开发了Microwell-seq,一种高通量,低成本的scRNA-seq平台。使用Microwell-seq,我们分析了覆盖所有主要小鼠器官的400,000多个单细胞,并构建了小鼠细胞图谱(MCA)的基本方案。我们揭示了以前没有很好表征的许多组织的单细胞层次结构。我们建立了一个基于Web的“单细胞MCA分析”管道,该管道可根据单细胞数字表达准确定义细胞类型。我们的研究证明了微孔测序技术和MCA资源的广泛适用性。















































 京公网安备 11010802027423号
京公网安备 11010802027423号