Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Modeling RNA-Binding Protein Specificity In Vivo by Precisely Registering Protein-RNA Crosslink Sites.
Molecular Cell ( IF 14.5 ) Pub Date : 2019-06-20 , DOI: 10.1016/j.molcel.2019.02.002 Huijuan Feng 1 , Suying Bao 1 , Mohammad Alinoor Rahman 2 , Sebastien M Weyn-Vanhentenryck 1 , Aziz Khan 3 , Justin Wong 1 , Ankeeta Shah 1 , Elise D Flynn 1 , Adrian R Krainer 2 , Chaolin Zhang 1
Molecular Cell ( IF 14.5 ) Pub Date : 2019-06-20 , DOI: 10.1016/j.molcel.2019.02.002 Huijuan Feng 1 , Suying Bao 1 , Mohammad Alinoor Rahman 2 , Sebastien M Weyn-Vanhentenryck 1 , Aziz Khan 3 , Justin Wong 1 , Ankeeta Shah 1 , Elise D Flynn 1 , Adrian R Krainer 2 , Chaolin Zhang 1
Affiliation
|
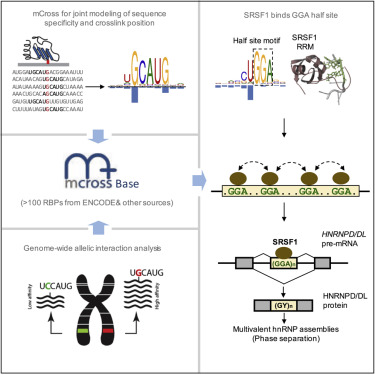
RNA-binding proteins (RBPs) regulate post-transcriptional gene expression by recognizing short and degenerate sequence motifs in their target transcripts, but precisely defining their binding specificity remains challenging. Crosslinking and immunoprecipitation (CLIP) allows for mapping of the exact protein-RNA crosslink sites, which frequently reside at specific positions in RBP motifs at single-nucleotide resolution. Here, we have developed a computational method, named mCross, to jointly model RBP binding specificity while precisely registering the crosslinking position in motif sites. We applied mCross to 112 RBPs using ENCODE eCLIP data and validated the reliability of the discovered motifs by genome-wide analysis of allelic binding sites. Our analyses revealed that the prototypical SR protein SRSF1 recognizes clusters of GGA half-sites in addition to its canonical GGAGGA motif. Therefore, SRSF1 regulates splicing of a much larger repertoire of transcripts than previously appreciated, including HNRNPD and HNRNPDL, which are involved in multivalent protein assemblies and phase separation.
中文翻译:

通过精确注册蛋白质-RNA 交联位点来模拟体内 RNA 结合蛋白质特异性。
RNA结合蛋白(RBP)通过识别目标转录物中的短和简并序列基序来调节转录后基因表达,但精确定义其结合特异性仍然具有挑战性。交联和免疫沉淀 (CLIP) 可以绘制精确的蛋白质-RNA 交联位点图谱,这些位点通常以单核苷酸分辨率位于 RBP 基序中的特定位置。在这里,我们开发了一种名为 mCross 的计算方法,用于联合模拟 RBP 结合特异性,同时精确记录基序位点中的交联位置。我们使用 ENCODE eCLIP 数据将 mCross 应用于 112 个 RBP,并通过等位基因结合位点的全基因组分析来验证所发现基序的可靠性。我们的分析表明,原型 SR 蛋白 SRSF1 除了其典型的 GGAGGA 基序外,还识别 GGA 半位点簇。因此,SRSF1 调节的转录本的剪接比之前认识的要大得多,包括 HNRNPD 和 HNRNPDL,它们参与多价蛋白组装和相分离。
更新日期:2019-06-20
中文翻译:

通过精确注册蛋白质-RNA 交联位点来模拟体内 RNA 结合蛋白质特异性。
RNA结合蛋白(RBP)通过识别目标转录物中的短和简并序列基序来调节转录后基因表达,但精确定义其结合特异性仍然具有挑战性。交联和免疫沉淀 (CLIP) 可以绘制精确的蛋白质-RNA 交联位点图谱,这些位点通常以单核苷酸分辨率位于 RBP 基序中的特定位置。在这里,我们开发了一种名为 mCross 的计算方法,用于联合模拟 RBP 结合特异性,同时精确记录基序位点中的交联位置。我们使用 ENCODE eCLIP 数据将 mCross 应用于 112 个 RBP,并通过等位基因结合位点的全基因组分析来验证所发现基序的可靠性。我们的分析表明,原型 SR 蛋白 SRSF1 除了其典型的 GGAGGA 基序外,还识别 GGA 半位点簇。因此,SRSF1 调节的转录本的剪接比之前认识的要大得多,包括 HNRNPD 和 HNRNPDL,它们参与多价蛋白组装和相分离。































 京公网安备 11010802027423号
京公网安备 11010802027423号