当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Next-Generation Glycan Microarray Enabled by DNA-Coded Glycan Library and Next-Generation Sequencing Technology.
Analytical Chemistry ( IF 6.7 ) Pub Date : 2019-06-12 00:00:00 , DOI: 10.1021/acs.analchem.9b01988 Maomao Yan , Yuyang Zhu , Xueyun Liu , Yi Lasanajak , Jinglin Xiong , Jingqiao Lu , Xi Lin , David Ashline 1 , Vernon Reinhold 1 , David F Smith , Xuezheng Song
Analytical Chemistry ( IF 6.7 ) Pub Date : 2019-06-12 00:00:00 , DOI: 10.1021/acs.analchem.9b01988 Maomao Yan , Yuyang Zhu , Xueyun Liu , Yi Lasanajak , Jinglin Xiong , Jingqiao Lu , Xi Lin , David Ashline 1 , Vernon Reinhold 1 , David F Smith , Xuezheng Song
Affiliation
|
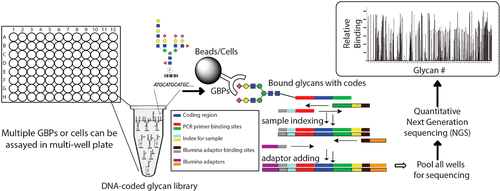
Interactions of glycans with proteins, cells, and microorganisms play important roles in cell–cell adhesion and host–pathogen interaction. Glycan microarray technology, in which multiple glycan structures are immobilized on a single glass slide and interrogated with glycan-binding proteins (GBPs), has become an indispensable tool in the study of protein–glycan interactions. Despite its great success, the current format of the glycan microarray requires expensive, specialized instrumentation and labor-intensive assay and image processing procedures, which limit automation and possibilities for high-throughput analyses. Furthermore, the current microarray is not suitable for assaying interaction with intact cells due to their large size compared to the two-dimensional microarray surface. To address these limitations, we developed the next-generation glycan microarray (NGGM) based on artificial DNA coding of glycan structures. In this novel approach, a glycan library is presented as a mixture of glycans and glycoconjugates, each of which is coded with a unique oligonucleotide sequence (code). The glycan mixture is interrogated by GBPs followed by the separation of unbound coded glycans. The DNA sequences that identify individual bound glycans are quantitatively sequenced (decoded) by powerful next-generation sequencing (NGS) technology, and copied numbers of the DNA codes represent relative binding specificities of corresponding glycan structures to GBPs. We demonstrate that NGGM generates glycan–GBP binding data that are consistent with that generated in a slide-based glycan microarray. More importantly, the solution phase binding assay is directly applicable to identifying glycan binding to intact cells, which is often challenging using glass slide-based glycan microarrays.
中文翻译:

DNA编码的糖链文库和下一代测序技术支持的下一代糖链微阵列。
聚糖与蛋白质,细胞和微生物的相互作用在细胞与细胞的粘附以及宿主与病原体的相互作用中起着重要的作用。聚糖微阵列技术将多个聚糖结构固定在单个载玻片上,并与聚糖结合蛋白(GBP)进行询问,该技术已成为研究蛋白质-聚糖相互作用的必不可少的工具。尽管取得了巨大的成功,但目前的聚糖微阵列格式仍需要昂贵的专用仪器以及劳动强度大的测定和图像处理程序,这限制了自动化和高通量分析的可能性。此外,由于与二维微阵列表面相比其尺寸大,当前的微阵列不适合用于测定与完整细胞的相互作用。为了解决这些限制,我们开发了基于聚糖结构的人工DNA编码的下一代聚糖微阵列(NGGM)。在这种新颖的方法中,聚糖文库以聚糖和糖缀合物的混合物的形式呈现,它们各自用唯一的寡核苷酸序列(代码)编码。聚糖混合物被GBPs查询,然后分离未结合的编码聚糖。通过强大的下一代测序(NGS)技术对识别单个结合聚糖的DNA序列进行定量测序(解码),DNA编码的复制数代表相应聚糖结构与GBPs的相对结合特异性。我们证明了NGGM生成的聚糖-GBP结合数据与基于幻灯片的聚糖微阵列中产生的数据一致。更重要的是,
更新日期:2019-06-12
中文翻译:

DNA编码的糖链文库和下一代测序技术支持的下一代糖链微阵列。
聚糖与蛋白质,细胞和微生物的相互作用在细胞与细胞的粘附以及宿主与病原体的相互作用中起着重要的作用。聚糖微阵列技术将多个聚糖结构固定在单个载玻片上,并与聚糖结合蛋白(GBP)进行询问,该技术已成为研究蛋白质-聚糖相互作用的必不可少的工具。尽管取得了巨大的成功,但目前的聚糖微阵列格式仍需要昂贵的专用仪器以及劳动强度大的测定和图像处理程序,这限制了自动化和高通量分析的可能性。此外,由于与二维微阵列表面相比其尺寸大,当前的微阵列不适合用于测定与完整细胞的相互作用。为了解决这些限制,我们开发了基于聚糖结构的人工DNA编码的下一代聚糖微阵列(NGGM)。在这种新颖的方法中,聚糖文库以聚糖和糖缀合物的混合物的形式呈现,它们各自用唯一的寡核苷酸序列(代码)编码。聚糖混合物被GBPs查询,然后分离未结合的编码聚糖。通过强大的下一代测序(NGS)技术对识别单个结合聚糖的DNA序列进行定量测序(解码),DNA编码的复制数代表相应聚糖结构与GBPs的相对结合特异性。我们证明了NGGM生成的聚糖-GBP结合数据与基于幻灯片的聚糖微阵列中产生的数据一致。更重要的是,


















































 京公网安备 11010802027423号
京公网安备 11010802027423号