Nature Communications ( IF 14.7 ) Pub Date : 2019-06-03 , DOI: 10.1038/s41467-019-10368-w Xiao Fan 1, 2 , Jia Wang 1 , Xing Zhang 1 , Zi Yang 1, 2 , Jin-Can Zhang 3 , Lingyun Zhao 1 , Hai-Lin Peng 3 , Jianlin Lei 1, 2 , Hong-Wei Wang 1, 2
|
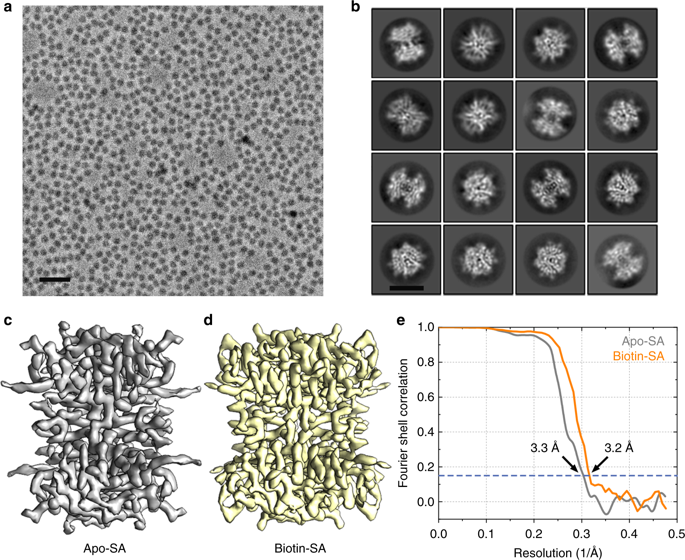
The fast development of single-particle cryogenic electron microscopy (cryo-EM) has made it more feasible to obtain the 3D structure of well-behaved macromolecules with a molecular weight higher than 300 kDa at ~3 Å resolution. However, it remains a challenge to obtain the high-resolution structures of molecules smaller than 200 kDa using single-particle cryo-EM. In this work, we apply the Cs-corrector-VPP-coupled cryo-EM to study the 52 kDa streptavidin (SA) protein supported on a thin layer of graphene and embedded in vitreous ice. We are able to solve both the apo-SA and biotin-bound SA structures at near-atomic resolution using single-particle cryo-EM. We demonstrate that the method has the potential to determine the structures of molecules as small as 39 kDa.
中文翻译:

52 kDa链霉抗生物素蛋白在3.2埃分辨率下的单粒子冷冻EM重建。
单粒子低温电子显微镜(cryo-EM)的快速发展使以3Å分辨率获得分子量高于300 kDa的行为良好的大分子的3D结构变得更加可行。然而,使用单粒子冷冻EM获得小于200 kDa的分子的高分辨率结构仍然是一个挑战。在这项工作中,我们应用Cs-校正剂-VPP耦合的冷冻电磁场研究了支撑在石墨烯薄层上并嵌入玻璃冰中的52 kDa链霉亲和素(SA)蛋白。我们能够使用单粒子冷冻电磁场以接近原子的分辨率解决apo-SA和生物素结合的SA结构。我们证明该方法具有确定小至39 kDa分子结构的潜力。





















































 京公网安备 11010802027423号
京公网安备 11010802027423号