Nature Communications ( IF 14.7 ) Pub Date : 2019-05-30 , DOI: 10.1038/s41467-019-10007-4 Florian Levet 1, 2, 3, 4, 5 , Guillaume Julien 1, 2 , Rémi Galland 1, 2 , Corey Butler 1, 2 , Anne Beghin 1, 2 , Anaël Chazeau 1, 2 , Philipp Hoess 6 , Jonas Ries 6 , Grégory Giannone 1, 2 , Jean-Baptiste Sibarita 1, 2
|
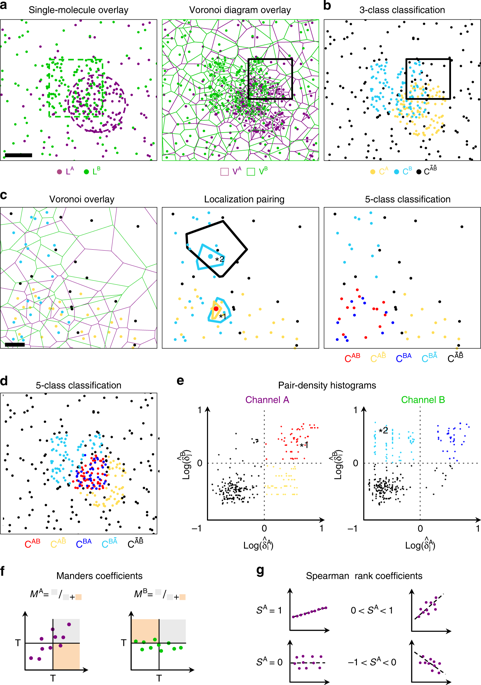
Multicolor single-molecule localization microscopy (λSMLM) is a powerful technique to reveal the relative nanoscale organization and potential colocalization between different molecular species. While several standard analysis methods exist for pixel-based images, λSMLM still lacks such a standard. Moreover, existing methods only work on 2D data and are usually sensitive to the relative molecular organization, a very important parameter to consider in quantitative SMLM. Here, we present an efficient, parameter-free colocalization analysis method for 2D and 3D λSMLM using tessellation analysis. We demonstrate that our method allows for the efficient computation of several popular colocalization estimators directly from molecular coordinates and illustrate its capability to analyze multicolor SMLM data in a robust and efficient manner.
中文翻译:

基于镶嵌的共定位分析方法,用于单分子定位显微镜。
多色单分子定位显微镜(λSMLM)是一种强大的技术,可揭示不同分子种类之间的相对纳米级组织和潜在的共定位。尽管存在几种基于像素的图像的标准分析方法,但λSMLM仍缺乏这种标准。此外,现有方法仅适用于2D数据,并且通常对相对分子组织敏感,这是定量SMLM中要考虑的非常重要的参数。在这里,我们提出了一种使用细分分析的2D和3DλSMLM高效,无参数共定位分析方法。我们证明了我们的方法可以直接从分子坐标中高效地计算几种流行的共定位估计值,并说明其以健壮和有效的方式分析多色SMLM数据的能力。





















































 京公网安备 11010802027423号
京公网安备 11010802027423号