当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mapping connections in signaling networks with ambiguous modularity.
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-05-23 , DOI: 10.1038/s41540-019-0096-1
Daniel Lill 1, 2 , Oleksii S Rukhlenko 2 , Anthony James Mc Elwee 2 , Eugene Kashdan 2, 3 , Jens Timmer 1, 4 , Boris N Kholodenko 2, 5, 6, 7
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-05-23 , DOI: 10.1038/s41540-019-0096-1
Daniel Lill 1, 2 , Oleksii S Rukhlenko 2 , Anthony James Mc Elwee 2 , Eugene Kashdan 2, 3 , Jens Timmer 1, 4 , Boris N Kholodenko 2, 5, 6, 7
Affiliation
|
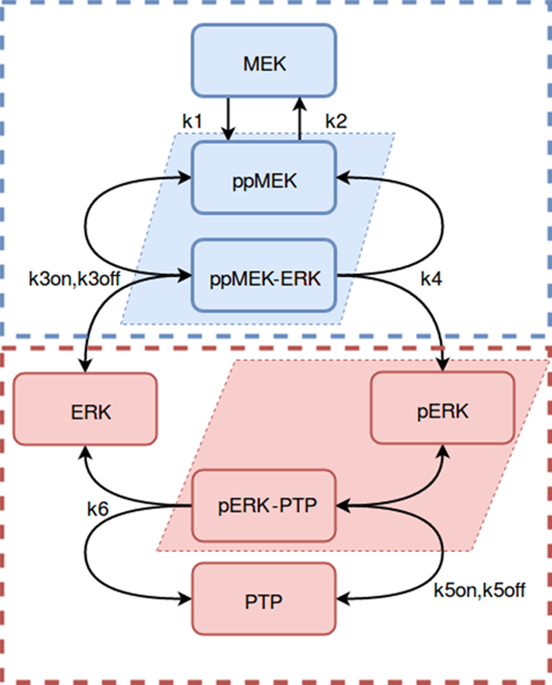
Modular Response Analysis (MRA) is a suite of methods that under certain assumptions permits the precise reconstruction of both the directions and strengths of connections between network modules from network responses to perturbations. Standard MRA assumes that modules are insulated, thereby neglecting the existence of inter-modular protein complexes. Such complexes sequester proteins from different modules and propagate perturbations to the protein abundance of a downstream module retroactively to an upstream module. MRA-based network reconstruction detects retroactive, sequestration-induced connections when an enzyme from one module is substantially sequestered by its substrate that belongs to a different module. Moreover, inferred networks may surprisingly depend on the choice of protein abundances that are experimentally perturbed, and also some inferred connections might be false. Here, we extend MRA by introducing a combined computational and experimental approach, which allows for a computational restoration of modular insulation, unmistakable network reconstruction and discrimination between solely regulatory and sequestration-induced connections for a range of signaling pathways. Although not universal, our approach extends MRA methods to signaling networks with retroactive interactions between modules arising from enzyme sequestration effects.
中文翻译:

映射具有不明确模块化的信令网络中的连接。
模块化响应分析(MRA)是一套方法,在某些假设下,可以精确重建网络模块之间从网络响应到扰动的连接方向和强度。标准 MRA 假设模块是绝缘的,从而忽略了模块间蛋白质复合物的存在。这种复合物隔离来自不同模块的蛋白质,并将对下游模块的蛋白质丰度的扰动传播到上游模块。当来自一个模块的酶被属于不同模块的底物充分隔离时,基于 MRA 的网络重建可检测追溯性隔离诱导的连接。此外,推断的网络可能令人惊讶地取决于实验扰动的蛋白质丰度的选择,并且一些推断的连接可能是错误的。在这里,我们通过引入组合的计算和实验方法来扩展 MRA,该方法允许对模块化绝缘进行计算恢复、明确无误的网络重建以及区分一系列信号通路的单独调节和隔离诱导的连接。尽管不通用,但我们的方法将 MRA 方法扩展到信号网络,并通过酶隔离效应产生的模块之间的追溯相互作用。
更新日期:2019-05-23
中文翻译:

映射具有不明确模块化的信令网络中的连接。
模块化响应分析(MRA)是一套方法,在某些假设下,可以精确重建网络模块之间从网络响应到扰动的连接方向和强度。标准 MRA 假设模块是绝缘的,从而忽略了模块间蛋白质复合物的存在。这种复合物隔离来自不同模块的蛋白质,并将对下游模块的蛋白质丰度的扰动传播到上游模块。当来自一个模块的酶被属于不同模块的底物充分隔离时,基于 MRA 的网络重建可检测追溯性隔离诱导的连接。此外,推断的网络可能令人惊讶地取决于实验扰动的蛋白质丰度的选择,并且一些推断的连接可能是错误的。在这里,我们通过引入组合的计算和实验方法来扩展 MRA,该方法允许对模块化绝缘进行计算恢复、明确无误的网络重建以及区分一系列信号通路的单独调节和隔离诱导的连接。尽管不通用,但我们的方法将 MRA 方法扩展到信号网络,并通过酶隔离效应产生的模块之间的追溯相互作用。




































 京公网安备 11010802027423号
京公网安备 11010802027423号