当前位置:
X-MOL 学术
›
Nat. Protoc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing.
Nature Protocols ( IF 13.1 ) Pub Date : 2019-05-03 , DOI: 10.1038/s41596-019-0159-1 Lionel A Sanz 1 , Frédéric Chédin 1
Nature Protocols ( IF 13.1 ) Pub Date : 2019-05-03 , DOI: 10.1038/s41596-019-0159-1 Lionel A Sanz 1 , Frédéric Chédin 1
Affiliation
|
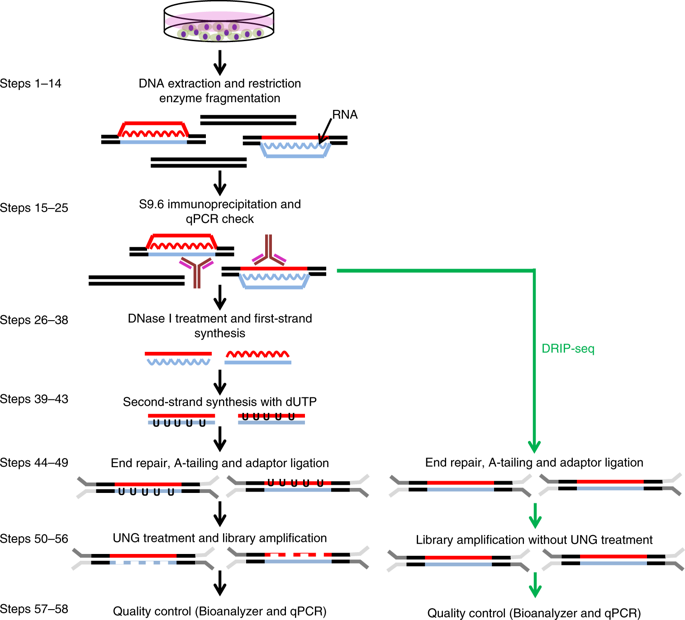
R-loops are prevalent three-stranded non-B DNA structures composed of an RNA-DNA hybrid and a single strand of DNA. R-loops are implicated in various basic nuclear processes, such as class-switch recombination, transcription termination and chromatin patterning. Perturbations in R-loop metabolism have been linked to genomic instability and have been implicated in human disorders, including cancer. As a consequence, the accurate mapping of these structures has been of increasing interest in recent years. Here, we describe two related immunoprecipitation-based methods for mapping R-loop structures: basic DRIP-seq (DNA-RNA immunoprecipitation followed by high-throughput DNA sequencing), an easy, robust, but resolution-limited technique; and DRIPc-seq (DNA-RNA immunoprecipitation followed by cDNA conversion coupled to high-throughput sequencing), a high-resolution and strand-specific iteration of the method that permits accurate R-loop mapping genome wide. Briefly, after gentle DNA extraction and restriction digestion with a cocktail of enzymes, R-loop structures are immunoprecipitated with the anti-RNA-DNA hybrid S9.6 antibody. Compared with DRIP-seq, in which the immunoprecipitated DNA is directly sequenced, DRIPc-seq permits the recovery of the RNA moiety of R-loops, and these RNA strands are subjected to strand-specific RNA sequencing (RNA-seq) analysis. DRIPc-seq can be performed in 5 d and can be applied to any cell type, provided sufficient starting material can be collected. Accurately mapping R-loop distribution in various cell lines and under varied conditions is essential to understanding the formation, roles and dynamic resolution of these important structures.
中文翻译:

通过基于S9.6的DNA-RNA免疫沉淀和高通量测序进行高分辨率,链特异性R环定位。
R环是由RNA-DNA杂合体和DNA的单链组成的普遍的三链非B DNA结构。R-环涉及各种基本的核过程,例如类别开关重组,转录终止和染色质图谱。R环代谢中的扰动已与基因组不稳定性相关联,并与包括癌症在内的人类疾病有关。结果,近年来,对这些结构的精确映射越来越引起关注。在这里,我们描述了两种相关的基于免疫沉淀的R环结构定位方法:基本的DRIP-seq(DNA-RNA免疫沉淀,然后进行高通量DNA测序),一种简单,稳健但分辨率受限的技术;和DRIPc-seq(DNA-RNA免疫沉淀,然后进行cDNA转换和高通量测序),该方法的高分辨率和链特异性迭代,可在全基因组范围内进行准确的R环定位。简而言之,在温和的DNA提取和酶混合物的限制性内切酶消化后,R-环结构被抗RNA-DNA杂合S9.6抗体免疫沉淀。与直接对免疫沉淀的DNA进行测序的DRIP-seq相比,DRIPc-seq可以回收R环的RNA部分,并对这些RNA链进行链特异性RNA测序(RNA-seq)分析。DRIPc-seq可以在5天内进行,并且可以应用于任何细胞类型,前提是可以收集到足够的起始原料。准确绘制各种细胞系中不同条件下的R环分布图对于理解这些重要结构的形成,作用和动态拆分至关重要。
更新日期:2019-05-16
中文翻译:

通过基于S9.6的DNA-RNA免疫沉淀和高通量测序进行高分辨率,链特异性R环定位。
R环是由RNA-DNA杂合体和DNA的单链组成的普遍的三链非B DNA结构。R-环涉及各种基本的核过程,例如类别开关重组,转录终止和染色质图谱。R环代谢中的扰动已与基因组不稳定性相关联,并与包括癌症在内的人类疾病有关。结果,近年来,对这些结构的精确映射越来越引起关注。在这里,我们描述了两种相关的基于免疫沉淀的R环结构定位方法:基本的DRIP-seq(DNA-RNA免疫沉淀,然后进行高通量DNA测序),一种简单,稳健但分辨率受限的技术;和DRIPc-seq(DNA-RNA免疫沉淀,然后进行cDNA转换和高通量测序),该方法的高分辨率和链特异性迭代,可在全基因组范围内进行准确的R环定位。简而言之,在温和的DNA提取和酶混合物的限制性内切酶消化后,R-环结构被抗RNA-DNA杂合S9.6抗体免疫沉淀。与直接对免疫沉淀的DNA进行测序的DRIP-seq相比,DRIPc-seq可以回收R环的RNA部分,并对这些RNA链进行链特异性RNA测序(RNA-seq)分析。DRIPc-seq可以在5天内进行,并且可以应用于任何细胞类型,前提是可以收集到足够的起始原料。准确绘制各种细胞系中不同条件下的R环分布图对于理解这些重要结构的形成,作用和动态拆分至关重要。































 京公网安备 11010802027423号
京公网安备 11010802027423号