The Lancet Psychiatry ( IF 30.8 ) Pub Date : 2019-04-15 , DOI: 10.1016/s2215-0366(19)30088-4
Marin M Jukic , Robert L Smith , Tore Haslemo , Espen Molden , Magnus Ingelman-Sundberg
|
Background
The polymorphic CYP2D6 enzyme metabolises the antipsychotic drugs risperidone and aripiprazole to their active metabolites, 9OH-risperidone and dehydroaripiprazole. The aim of this study was to quantify the effect of CYP2D6 genetic variability on risperidone and aripiprazole exposure and treatment in a large patient population.
Methods
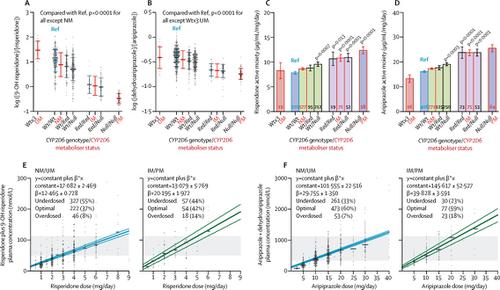
We retrospectively obtained patient data from a routine therapeutic drug monitoring database at the Center for Psychopharmacology, Diakonhjemmet Hospital, Oslo, Norway, between Jan 1, 2005, and Oct 15, 2018. Individuals included in our analyses were CYP2D6-genotyped patients treated with risperidone or aripiprazole. Inclusion criteria for measurement of pharmacokinetic parameters (drug and metabolite serum concentrations) were oral administration of risperidone or aripiprazole, information known about prescribed daily dose and comedications, and aged older than 18 years. Exclusion criteria included polypharmacy with drugs known to be CYP2D6 inhibitors or CYP3A4 inducers or inhibitors. Treatment failure was analysed in all patients treated with risperidone or aripiprazole without these criteria. The first endpoint in our analysis was the metabolism of risperidone to 9OH-risperidone and aripiprazole to dehydroaripiprazole, estimated by the log-transformed ratio between the concentrations of metabolite and parent drug (ie, the metabolic ratio for risperidone [9OH-risperidone]/[risperidone] and the metabolic ratio for aripiprazole [dehydroaripiprazole]/[aripiprazole]). Endpoint two was measurement of drug exposure, quantified by the dose-normalised sum of parent drug and active metabolite serum concentrations (ie, active moiety). The third endpoint of treatment failure was measured as the number of patients switched from risperidone or aripiprazole to another antipsychotic drug within 1 year after the last therapeutic drug monitoring analysis of risperidone or aripiprazole. Patient subgroups were defined by CYP2D6 genotype-determined metaboliser status: poor metabolisers, intermediate metabolisers, normal metabolisers, and ultrarapid metabolisers. ANOVA was used to assess the differences in metabolic ratios, active moieties, and daily doses between individual metaboliser categories, and risperidone and aripiprazole therapeutic failures were compared by logistic regression using the normal metaboliser subgroup as a reference.
Findings
1288 risperidone-treated patients and 1334 aripiprazole-treated patients were included in the study, of whom 725 (56%) risperidone-treated and 890 (67%) aripiprazole-treated patients were eligible for the pharmacokinetic analyses. CYP2D6 genotype significantly changed risperidone and aripiprazole metabolism resulting in an approximately 1·6-times and 1·4-times increase in risperidone and aripiprazole active moiety exposure in poor and intermediate metabolisers compared with normal metabolisers, respectively (odds ratios [OR] for the risperidone dose-normalised active moiety concentration 1·568, 95% CI 1·401–1·736, and 1·373, 1·213–1·532; and for the aripiprazole dose-normalised active moiety concentration 1·585, 1·447–1·724, and 1·476, 1·263–1·688, respectively; p<0·0001 for all). Compared with doses for normal metabolisers, clinicians reduced daily doses of risperidone and aripiprazole administered to poor metabolisers by 19% (95% CI 5–35, p=0·010) and 15% (95% CI 1–28, p=0·033) respectively. The incidence of switching from risperidone to another antipsychotic was increased in ultrarapid metabolisers (OR 2·934, 95% CI 1·437–5·989, p=0·003) and poor metabolisers (1·874, 1·128–3·112, p=0·015); by contrast, the incidence of switching from aripiprazole to another antipsychotic was not significantly related to CYP2D6 metaboliser status.
Interpretation
CYP2D6 genotype had a substantial clinical effect on risperidone and aripiprazole exposure and on the therapeutic failure of risperidone. Pre-emptive CYP2D6 genotyping would be valuable for individualising risperidone and aripiprazole dosing and treatment optimisation.
Funding
H2020 program U-PGx, The Swedish Research Council, the Swedish Brain foundation, and the South-Eastern Norway Regional Health Authority.
中文翻译:

的影响CYP2D6暴露和利培酮和阿立哌唑的功效基因型的回顾性队列研究
背景
多态性CYP2D6酶将抗精神病药利培酮和阿立哌唑代谢成其活性代谢产物9OH-利培酮和脱氢阿立哌唑。这项研究的目的是量化CYP2D6遗传变异对利培酮和阿立哌唑暴露和治疗的大量患者的影响。
方法
在2005年1月1日至2018年10月15日期间,我们从挪威奥斯陆Diakonhjemmet医院心理药理学中心的常规治疗药物监测数据库中回顾性收集了患者数据。我们纳入的分析对象为CYP2D6利培酮或阿立哌唑治疗的基因型患者。药代动力学参数(药物和代谢产物血清浓度)的测量纳入标准为口服利培酮或阿立哌唑,已知的每日处方剂量和喜剧信息以及年龄大于18岁。排除标准包括与已知为CYP2D6抑制剂或CYP3A4诱导剂或抑制剂的药物合用。在没有这些标准的情况下,对所有使用利培酮或阿立哌唑治疗的患者的治疗失败进行了分析。我们分析的第一个终点是利培酮至9OH-利培酮的代谢以及阿立哌唑至脱氢阿立哌唑的代谢,通过代谢物与母体药物的浓度之间的对数转换比来估算(即,(利培酮[9OH-利培酮] / [利培酮]的代谢率和阿立哌唑[dehydroaripiprazole] / [aripiprazole]的代谢率)。终点二是药物暴露的测量,通过母体药物和活性代谢物血清浓度(即活性部分)的剂量标准化总和进行量化。在对利培酮或阿立哌唑进行最后一次治疗药物监测分析后的一年内,从患者人数从利培酮或阿立哌唑转用另一种抗精神病药物的方法来衡量治疗失败的第三个终点。患者亚组定义为 在对利培酮或阿立哌唑进行最后一次治疗药物监测分析后的一年内,将患者人数从利培酮或阿立哌唑转换为另一种抗精神病药物的方法来衡量治疗失败的第三个终点。患者亚组定义为 在对利培酮或阿立哌唑进行最后一次治疗药物监测分析后的一年内,从患者人数从利培酮或阿立哌唑转用另一种抗精神病药物的方法来衡量治疗失败的第三个终点。患者亚组定义为CYP2D6基因型决定的代谢者状态:弱代谢者,中间代谢者,正常代谢者和超快速代谢者。方差分析用于评估各个代谢类别之间的代谢率,活性部分和日剂量之间的差异,利培酮和阿立哌唑的治疗失败率以正常代谢者亚组为参考,通过逻辑回归进行比较。
发现
这项研究包括1288例利培酮治疗的患者和1334例阿立哌唑治疗的患者,其中725(56%)利培酮治疗的患者和890(67%)阿立哌唑治疗的患者符合药代动力学分析的条件。CYP2D6基因型显着改变了利培酮和阿立哌唑的代谢,与正常代谢者相比,不良和中度代谢者的利培酮和阿立哌唑活性部分暴露分别增加了约1·6倍和1·4倍(利培酮的比值比[OR]剂量标准化的活性部分浓度1·568、95%CI 1·401-1·736和1·373、1·213-1·532;对于阿立哌唑,剂量标准化的活性部分浓度1·585、1·分别为447-1·724和1·476、1·263-1·688;对于全部p <0·0001)。与正常代谢者的剂量相比,临床医生减少了对不良代谢者服用利培酮和阿立哌唑的每日剂量,分别降低了19%(95%CI 5–35,p = 0·010)和15%(95%CI 1–28,p = 0) ·033)。超快代谢者(OR 2·934,95%CI 1·437-5·989,p = 0·003)和不良代谢者(1·874、1·128-3)从利培酮转换为另一种抗精神病药的发生率增加。 ·112,p = 0·015);相比之下,从阿立哌唑转换为另一种抗精神病药的发生率与CYP2D6代谢状态无关。
解释
CYP2D6基因型对利培酮和阿立哌唑的暴露以及利培酮的治疗失败具有重要的临床作用。CYP2D6的先发性基因分型对于利培酮和阿立哌唑剂量的个体化和治疗的优化将是有价值的。
资金
H2020计划U-PGx,瑞典研究委员会,瑞典大脑基金会和挪威东南部地区卫生局。































 京公网安备 11010802027423号
京公网安备 11010802027423号