当前位置:
X-MOL 学术
›
Mol. Syst. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
INKA, an integrative data analysis pipeline for phosphoproteomic inference of active kinases.
Molecular Systems Biology ( IF 8.5 ) Pub Date : 2019-04-12 , DOI: 10.15252/msb.20188250
Robin Beekhof 1, 2 , Carolien van Alphen 1, 2 , Alex A Henneman 1, 2 , Jaco C Knol 1, 2 , Thang V Pham 1, 2 , Frank Rolfs 1, 2 , Mariette Labots 1 , Evan Henneberry 2 , Tessa Ys Le Large 1, 2 , Richard R de Haas 1, 2 , Sander R Piersma 1, 2 , Valentina Vurchio 3 , Andrea Bertotti 3 , Livio Trusolino 3 , Henk Mw Verheul 1 , Connie R Jimenez 2, 4
Molecular Systems Biology ( IF 8.5 ) Pub Date : 2019-04-12 , DOI: 10.15252/msb.20188250
Robin Beekhof 1, 2 , Carolien van Alphen 1, 2 , Alex A Henneman 1, 2 , Jaco C Knol 1, 2 , Thang V Pham 1, 2 , Frank Rolfs 1, 2 , Mariette Labots 1 , Evan Henneberry 2 , Tessa Ys Le Large 1, 2 , Richard R de Haas 1, 2 , Sander R Piersma 1, 2 , Valentina Vurchio 3 , Andrea Bertotti 3 , Livio Trusolino 3 , Henk Mw Verheul 1 , Connie R Jimenez 2, 4
Affiliation
|
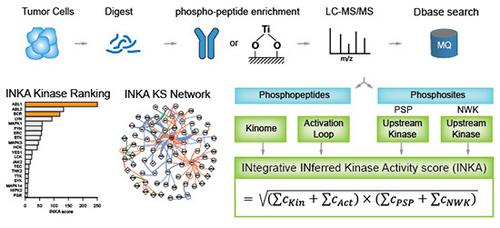
Identifying hyperactive kinases in cancer is crucial for individualized treatment with specific inhibitors. Kinase activity can be discerned from global protein phosphorylation profiles obtained with mass spectrometry-based phosphoproteomics. A major challenge is to relate such profiles to specific hyperactive kinases fueling growth/progression of individual tumors. Hitherto, the focus has been on phosphorylation of either kinases or their substrates. Here, we combined label-free kinase-centric and substrate-centric information in an Integrative Inferred Kinase Activity (INKA) analysis. This multipronged, stringent analysis enables ranking of kinase activity and visualization of kinase-substrate networks in a single biological sample. To demonstrate utility, we analyzed (i) cancer cell lines with known oncogenes, (ii) cell lines in a differential setting (wild-type versus mutant, +/- drug), (iii) pre- and on-treatment tumor needle biopsies, (iv) cancer cell panel with available drug sensitivity data, and (v) patient-derived tumor xenografts with INKA-guided drug selection and testing. These analyses show superior performance of INKA over its components and substrate-based single-sample tool KARP, and underscore target potential of high-ranking kinases, encouraging further exploration of INKA's functional and clinical value.
中文翻译:

INKA,用于活性激酶磷酸化蛋白质组学推断的综合数据分析管道。
识别癌症中过度活跃的激酶对于使用特定抑制剂进行个体化治疗至关重要。激酶活性可以通过基于质谱的磷酸化蛋白质组学获得的整体蛋白质磷酸化谱来辨别。一个主要的挑战是将这些特征与促进个体肿瘤生长/进展的特定高活性激酶联系起来。迄今为止,焦点一直集中在激酶或其底物的磷酸化上。在这里,我们在综合推断激酶活性 (INKA) 分析中结合了无标记的以激酶为中心和以底物为中心的信息。这种多管齐下、严格的分析可以对单个生物样本中的激酶活性进行排序并实现激酶-底物网络的可视化。为了证明实用性,我们分析了(i)具有已知癌基因的癌细胞系,(ii)不同环境下的细胞系(野生型与突变型,+/-药物),(iii)治疗前和治疗中的肿瘤针活检,(iv)具有可用药物敏感性数据的癌细胞组,以及(v)具有 INKA 指导药物选择和测试的患者来源的肿瘤异种移植物。这些分析显示 INKA 的性能优于其成分和基于底物的单样本工具 KARP,并强调了高级激酶的目标潜力,鼓励进一步探索 INKA 的功能和临床价值。
更新日期:2019-06-21
中文翻译:

INKA,用于活性激酶磷酸化蛋白质组学推断的综合数据分析管道。
识别癌症中过度活跃的激酶对于使用特定抑制剂进行个体化治疗至关重要。激酶活性可以通过基于质谱的磷酸化蛋白质组学获得的整体蛋白质磷酸化谱来辨别。一个主要的挑战是将这些特征与促进个体肿瘤生长/进展的特定高活性激酶联系起来。迄今为止,焦点一直集中在激酶或其底物的磷酸化上。在这里,我们在综合推断激酶活性 (INKA) 分析中结合了无标记的以激酶为中心和以底物为中心的信息。这种多管齐下、严格的分析可以对单个生物样本中的激酶活性进行排序并实现激酶-底物网络的可视化。为了证明实用性,我们分析了(i)具有已知癌基因的癌细胞系,(ii)不同环境下的细胞系(野生型与突变型,+/-药物),(iii)治疗前和治疗中的肿瘤针活检,(iv)具有可用药物敏感性数据的癌细胞组,以及(v)具有 INKA 指导药物选择和测试的患者来源的肿瘤异种移植物。这些分析显示 INKA 的性能优于其成分和基于底物的单样本工具 KARP,并强调了高级激酶的目标潜力,鼓励进一步探索 INKA 的功能和临床价值。



































 京公网安备 11010802027423号
京公网安备 11010802027423号