Cell Systems ( IF 9.0 ) Pub Date : 2019-04-04 , DOI: 10.1016/j.cels.2019.03.004 Joel Rozowsky 1 , Robert R Kitchen 1 , Jonathan J Park 1 , Timur R Galeev 1 , James Diao 2 , Jonathan Warrell 1 , William Thistlethwaite 3 , Sai L Subramanian 3 , Aleksandar Milosavljevic 3 , Mark Gerstein 4
|
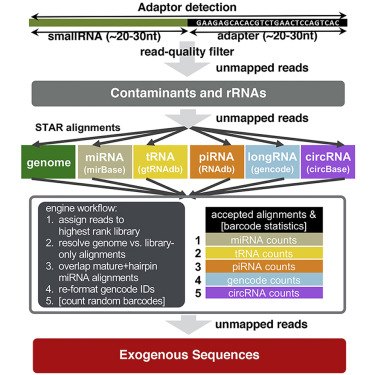
Small RNA sequencing has been widely adopted to study the diversity of extracellular RNAs (exRNAs) in biofluids; however, the analysis of exRNA samples can be challenging: they are vulnerable to contamination and artifacts from different isolation techniques, present in lower concentrations than cellular RNA, and occasionally of exogenous origin. To address these challenges, we present exceRpt, the exRNA-processing toolkit of the NIH Extracellular RNA Communication Consortium (ERCC). exceRpt is structured as a cascade of filters and quantifications prioritized based on one’s confidence in a given set of annotated RNAs. It generates quality control reports and abundance estimates for RNA biotypes. It is also capable of characterizing mappings to exogenous genomes, which, in turn, can be used to generate phylogenetic trees. exceRpt has been used to uniformly process all ∼3,500 exRNA-seq datasets in the public exRNA Atlas and is available from genboree.org and github.gersteinlab.org/exceRpt.
中文翻译:

摘录:用于细胞外RNA分析的综合分析平台。
小RNA测序已被广泛用于研究生物流体中细胞外RNA(exRNA)的多样性。然而,对exRNA样品的分析可能具有挑战性:它们容易受到来自不同分离技术的污染和伪影的影响,其浓度低于细胞RNA,并且有时是外源性的。为了解决这些挑战,我们介绍了exceRpt,NIH细胞外RNA通讯联盟(ERCC)的exRNA处理工具包。exceRpt结构为一连串的过滤器和定量分析,基于对一组给定注释RNA的置信度来确定优先级。它生成RNA生物型的质量控制报告和丰度估计。它也能够表征到外源基因组的定位,进而可用于生成系统发育树。































 京公网安备 11010802027423号
京公网安备 11010802027423号