当前位置:
X-MOL 学术
›
Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Determination of the Selection Capacity of Antibiotics for Gene Selection
Biotechnology Journal ( IF 3.2 ) Pub Date : 2018-02-13 , DOI: 10.1002/biot.201700747
Iris Delrue 1 , Qiubao Pan 2 , Anna K. Baczmanska 1 , Bram W. Callens 1 , Lia L. M. Verdoodt 3
Biotechnology Journal ( IF 3.2 ) Pub Date : 2018-02-13 , DOI: 10.1002/biot.201700747
Iris Delrue 1 , Qiubao Pan 2 , Anna K. Baczmanska 1 , Bram W. Callens 1 , Lia L. M. Verdoodt 3
Affiliation
|
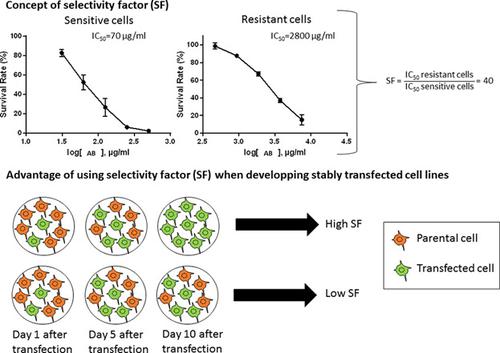
Choosing a potent selection antibiotic (SA), is a crucial success factor when creating stably transfected cell lines using an antibiotic selection marker. The selection capacity of this antibiotic is defined as its ability to kill sensitive, untransfected parental cells, while leaving resistant, transfected cells unharmed. Currently, no procedure has been described to determine this selection capacity. Therefore, a protocol to obtain a numerical value, called the “selectivity factor” (SF), that defines the selection capacity of SAs is developed. The SF is determined by using a modified MTT (3‐(4,5‐dimethylthiazol‐2‐yl)‐diphenyltetrazolium bromide) assay for both sensitive and resistant cells, and applies to commonly used cell lines. To prove the concept, the SF of the SA G418 and hygromycin B (HmB) on several cell lines is determined. The SF of G418 on BHK‐21 cells is very high, indicating that G418 is an ideal SA for transfected BHK‐21 cells. For HeLa cells, the SF of G418 is very low suggesting G418 is not an optimal SA for selecting transfected HeLa cells. For these cells, HmB would be a better choice. These conclusions are confirmed by an independent cell death assay. The SF identifies the most optimal SA for a certain cell line, reduces the risk of selecting spontaneously resistant cell clones, and streamlines the process of generating stable cell lines. Most importantly, the method is especially time saving when obtaining stable cell lines expressing toxic genes, and reduces culture times for generating large numbers of cell lines from the same parental cell line.
中文翻译:

确定抗生素选择基因的能力
当使用抗生素选择标记创建稳定转染的细胞系时,选择有效的选择抗生素(SA)是至关重要的成功因素。该抗生素的选择能力被定义为其杀死敏感的,未转染的亲代细胞,而使抗性,转染的细胞不受伤害的能力。当前,没有描述确定该选择能力的过程。因此,开发了一种用于获取数值的协议,该协议称为“选择性因子”(SF),该协议定义了SA的选择能力。通过使用改良的MTT(3-(4,5-二甲基噻唑-2-基)-溴化二苯基四唑鎓)测定法测定敏感和耐药细胞的SF,并适用于常用细胞系。为了证明这一概念,确定了SA G418和潮霉素B(HmB)在几种细胞系中的SF。B418-21细胞上G418的SF很高,表明G418是转染的BHK-21细胞的理想SA。对于HeLa细胞,G418的SF非常低,表明G418不是选择转染HeLa细胞的最佳SA。对于这些细胞,HmB将是更好的选择。通过独立的细胞死亡测定法证实了这些结论。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。对于HeLa细胞,G418的SF非常低,表明G418不是选择转染HeLa细胞的最佳SA。对于这些细胞,HmB将是更好的选择。通过独立的细胞死亡测定法证实了这些结论。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。对于HeLa细胞,G418的SF非常低,表明G418不是选择转染HeLa细胞的最佳SA。对于这些细胞,HmB将是更好的选择。通过独立的细胞死亡测定法证实了这些结论。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。
更新日期:2018-06-03
中文翻译:

确定抗生素选择基因的能力
当使用抗生素选择标记创建稳定转染的细胞系时,选择有效的选择抗生素(SA)是至关重要的成功因素。该抗生素的选择能力被定义为其杀死敏感的,未转染的亲代细胞,而使抗性,转染的细胞不受伤害的能力。当前,没有描述确定该选择能力的过程。因此,开发了一种用于获取数值的协议,该协议称为“选择性因子”(SF),该协议定义了SA的选择能力。通过使用改良的MTT(3-(4,5-二甲基噻唑-2-基)-溴化二苯基四唑鎓)测定法测定敏感和耐药细胞的SF,并适用于常用细胞系。为了证明这一概念,确定了SA G418和潮霉素B(HmB)在几种细胞系中的SF。B418-21细胞上G418的SF很高,表明G418是转染的BHK-21细胞的理想SA。对于HeLa细胞,G418的SF非常低,表明G418不是选择转染HeLa细胞的最佳SA。对于这些细胞,HmB将是更好的选择。通过独立的细胞死亡测定法证实了这些结论。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。对于HeLa细胞,G418的SF非常低,表明G418不是选择转染HeLa细胞的最佳SA。对于这些细胞,HmB将是更好的选择。通过独立的细胞死亡测定法证实了这些结论。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。对于HeLa细胞,G418的SF非常低,表明G418不是选择转染HeLa细胞的最佳SA。对于这些细胞,HmB将是更好的选择。通过独立的细胞死亡测定法证实了这些结论。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。SF为特定细胞系确定了最理想的SA,降低了选择自发抗性细胞克隆的风险,并简化了生成稳定细胞系的过程。最重要的是,当获得表达毒性基因的稳定细胞系时,该方法特别节省时间,并且减少了用于从同一亲代细胞系生成大量细胞系的培养时间。

































 京公网安备 11010802027423号
京公网安备 11010802027423号