Nature Communications ( IF 14.7 ) Pub Date : 2019-03-27 , DOI: 10.1038/s41467-019-09369-6 Xiuhai Mao , Ke Li , Mengmeng Liu , Xinyu Wang , Tianxin Zhao , Bolin An , Mengkui Cui , Yingfeng Li , Jiahua Pu , Jiang Li , Lihua Wang , Timothy K. Lu , Chunhai Fan , Chao Zhong
|
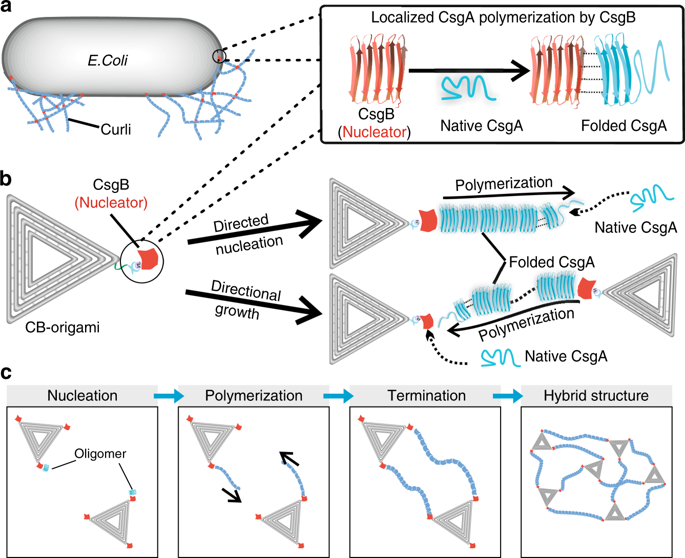
The physiological or pathological formation of fibrils often relies on molecular-scale nucleators that finely control the kinetics and structural features. However, mechanistic understanding of how protein nucleators mediate fibril formation in cells remains elusive. Here, we develop a CsgB-decorated DNA origami (CB-origami) to mimic protein nucleators in Escherichia coli biofilm that direct curli polymerization. We show that CB-origami directs curli subunit CsgA monomers to form oligomers and then accelerates fibril formation by increasing the proliferation rate of primary pathways. Fibrils grow either out from (departure mode) or towards the nucleators (arrival mode), implying two distinct roles of CsgB: as nucleation sites and as trap sites to capture growing nanofibrils in vicinity. Curli polymerization follows typical stop-and-go dynamics but exhibits a higher instantaneous elongation rate compared with independent fibril growth. This origami nucleator thus provides an in vitro platform for mechanistically probing molecular nucleation and controlling directional fibril polymerization for bionanotechnology.
中文翻译:

用DNA折纸成核剂指导curli聚合
原纤维的生理或病理学形成通常依赖于精确控制动力学和结构特征的分子级成核剂。然而,对蛋白质成核剂如何介导细胞中原纤维形成的机械理解仍然难以捉摸。在这里,我们开发了CsgB装饰的DNA折纸(CB-origami),以模拟大肠杆菌中的蛋白成核剂指导curli聚合的生物膜。我们表明,CB折纸指导curli亚基CsgA单体形成低聚物,然后通过增加主要途径的增殖速率来加速原纤维形成。原纤维要么从(离开模式)生长出来,要么向成核器(到达模式)生长,这暗示着CsgB的两个不同作用:作为成核位点和作为捕获位点以捕获附近生长的纳米原纤维。卷曲聚合遵循典型的走走停停动力学,但与独立的原纤维生长相比,表现出更高的瞬时伸长率。因此,该折纸成核器提供了一个体外平台,用于机械探测分子成核并控制用于仿生纳米技术的定向原纤维聚合。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号