Scientific Reports ( IF 3.8 ) Pub Date : 2019-03-18 , DOI: 10.1038/s41598-019-41215-z Quan Peng , Chang Xu , Daniel Kim , Marcus Lewis , John DiCarlo , Yexun Wang
|
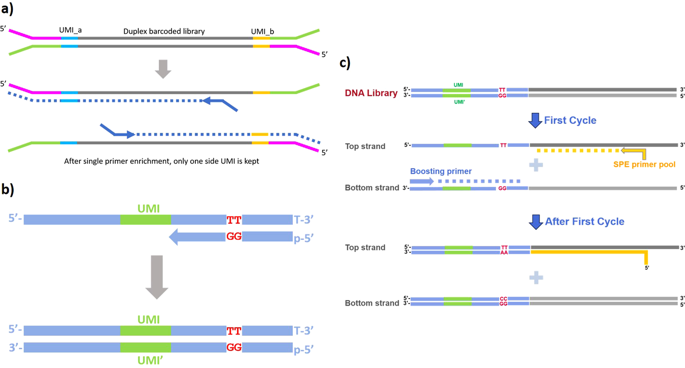
For specific detection of somatic variants at very low levels, artifacts from the NGS workflow have to be eliminated. Various approaches using unique molecular identifiers (UMI) to analytically remove NGS artifacts have been described. Among them, Duplex-seq was shown to be highly effective, by leveraging the sequence complementarity of two DNA strands. However, all of the published Duplex-seq implementations so far required pair-end sequencing and in the case of combining duplex sequencing with target enrichment, lengthy hybridization enrichment was required. We developed a simple protocol, which enabled the retrieval of duplex UMI in multiplex PCR based enrichment and sequencing. Using this protocol and reference materials, we demonstrated the accurate detection of known SNVs at 0.1–0.2% allele fractions, aided by duplex UMI. We also observed that low level base substitution artifacts could be introduced when preparing in vitro DNA reference materials, which could limit their utility as a benchmarking tool for variant detection at very low levels. Our new targeted sequencing method offers the benefit of using duplex UMI to remove NGS artifacts in a much more simplified workflow than existing targeted duplex sequencing methods.
中文翻译:

单端双链-UMI的靶向单引物富集测序
为了特异地检测极低水平的体细胞变异,必须消除NGS工作流程中的假象。已经描述了使用唯一分子标识符(UMI)来分析地去除NGS伪像的各种方法。其中,通过利用两条DNA链的序列互补性,Duplex-seq被证明是高效的。但是,到目前为止,所有已发布的Duplex-seq实施都需要成对测序,并且在将双链测序与靶标富集结合的情况下,需要长时间的杂交富集。我们开发了一种简单的协议,该协议使得能够在基于多重PCR的富集和测序中检索双工UMI。使用该方案和参考资料,我们证明了在双联UMI的辅助下,可以以0.1–0.2%等位基因分数准确检测已知的SNV。体外DNA参考材料,这可能限制了它们作为非常低水平的变体检测的基准工具的实用性。我们的新型靶向测序方法的优势在于,与现有的靶向双链测序方法相比,使用双工UMI可以以更加简化的工作流程去除NGS伪像。































 京公网安备 11010802027423号
京公网安备 11010802027423号