Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structural basis for DNMT3A-mediated de novo DNA methylation
Nature ( IF 50.5 ) Pub Date : 2018-02-01 , DOI: 10.1038/nature25477 Zhi-Min Zhang , Rui Lu , Pengcheng Wang , Yang Yu , Dongliang Chen , Linfeng Gao , Shuo Liu , Debin Ji , Scott B Rothbart , Yinsheng Wang , Gang Greg Wang , Jikui Song
Nature ( IF 50.5 ) Pub Date : 2018-02-01 , DOI: 10.1038/nature25477 Zhi-Min Zhang , Rui Lu , Pengcheng Wang , Yang Yu , Dongliang Chen , Linfeng Gao , Shuo Liu , Debin Ji , Scott B Rothbart , Yinsheng Wang , Gang Greg Wang , Jikui Song
|
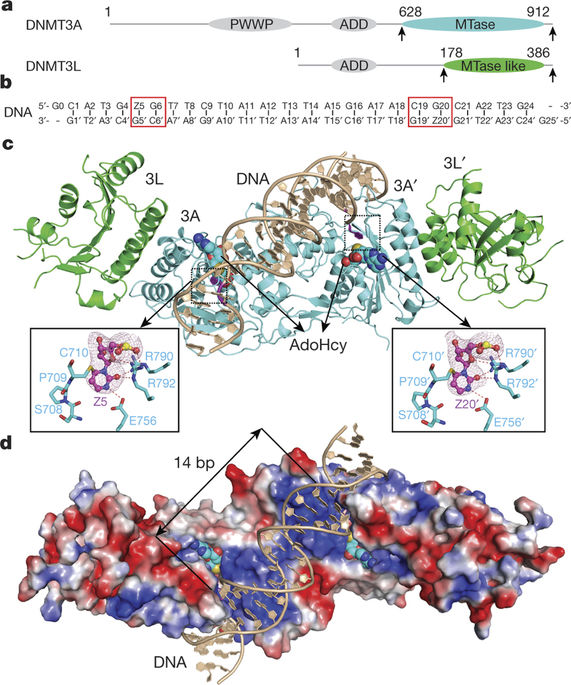
DNA methylation by de novo DNA methyltransferases 3A (DNMT3A) and 3B (DNMT3B) at cytosines is essential for genome regulation and development. Dysregulation of this process is implicated in various diseases, notably cancer. However, the mechanisms underlying DNMT3 substrate recognition and enzymatic specificity remain elusive. Here we report a 2.65-ångström crystal structure of the DNMT3A–DNMT3L–DNA complex in which two DNMT3A monomers simultaneously attack two cytosine–phosphate–guanine (CpG) dinucleotides, with the target sites separated by 14 base pairs within the same DNA duplex. The DNMT3A–DNA interaction involves a target recognition domain, a catalytic loop, and DNMT3A homodimeric interface. Arg836 of the target recognition domain makes crucial contacts with CpG, ensuring DNMT3A enzymatic preference towards CpG sites in cells. Haematological cancer-associated somatic mutations of the substrate-binding residues decrease DNMT3A activity, induce CpG hypomethylation, and promote transformation of haematopoietic cells. Together, our study reveals the mechanistic basis for DNMT3A-mediated DNA methylation and establishes its aetiological link to human disease.
中文翻译:

DNMT3A 介导的从头 DNA 甲基化的结构基础
胞嘧啶从头 DNA 甲基转移酶 3A (DNMT3A) 和 3B (DNMT3B) 引起的 DNA 甲基化对于基因组调控和发育至关重要。这个过程的失调与各种疾病有关,尤其是癌症。然而,DNMT3 底物识别和酶特异性的潜在机制仍然难以捉摸。在这里,我们报告了 DNMT3A-DNMT3L-DNA 复合物的 2.65-ångström 晶体结构,其中两个 DNMT3A 单体同时攻击两个胞嘧啶-磷酸-鸟嘌呤 (CpG) 二核苷酸,目标位点在同一 DNA 双链体中被 14 个碱基对隔开。DNMT3A-DNA 相互作用涉及目标识别域、催化环和 DNMT3A 同源二聚体界面。目标识别域的 Arg836 与 CpG 进行关键接触,确保 DNMT3A 对细胞中 CpG 位点的酶促偏好。底物结合残基的血液学癌症相关体细胞突变降低 DNMT3A 活性,诱导 CpG 低甲基化,并促进造血细胞的转化。总之,我们的研究揭示了 DNMT3A 介导的 DNA 甲基化的机制基础,并建立了其与人类疾病的病因学联系。
更新日期:2018-02-01
中文翻译:

DNMT3A 介导的从头 DNA 甲基化的结构基础
胞嘧啶从头 DNA 甲基转移酶 3A (DNMT3A) 和 3B (DNMT3B) 引起的 DNA 甲基化对于基因组调控和发育至关重要。这个过程的失调与各种疾病有关,尤其是癌症。然而,DNMT3 底物识别和酶特异性的潜在机制仍然难以捉摸。在这里,我们报告了 DNMT3A-DNMT3L-DNA 复合物的 2.65-ångström 晶体结构,其中两个 DNMT3A 单体同时攻击两个胞嘧啶-磷酸-鸟嘌呤 (CpG) 二核苷酸,目标位点在同一 DNA 双链体中被 14 个碱基对隔开。DNMT3A-DNA 相互作用涉及目标识别域、催化环和 DNMT3A 同源二聚体界面。目标识别域的 Arg836 与 CpG 进行关键接触,确保 DNMT3A 对细胞中 CpG 位点的酶促偏好。底物结合残基的血液学癌症相关体细胞突变降低 DNMT3A 活性,诱导 CpG 低甲基化,并促进造血细胞的转化。总之,我们的研究揭示了 DNMT3A 介导的 DNA 甲基化的机制基础,并建立了其与人类疾病的病因学联系。































 京公网安备 11010802027423号
京公网安备 11010802027423号