当前位置:
X-MOL 学术
›
Nat. Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
FTO controls reversible m6Am RNA methylation during snRNA biogenesis.
Nature Chemical Biology ( IF 12.9 ) Pub Date : 2019-02-18 , DOI: 10.1038/s41589-019-0231-8 Jan Mauer 1, 2 , Miriam Sindelar 1 , Vladimir Despic 1 , Théo Guez 3 , Ben R Hawley 1 , Jean-Jacques Vasseur 3 , Andrea Rentmeister 4 , Steven S Gross 1 , Livio Pellizzoni 5 , Françoise Debart 3 , Hani Goodarzi 6 , Samie R Jaffrey 1
Nature Chemical Biology ( IF 12.9 ) Pub Date : 2019-02-18 , DOI: 10.1038/s41589-019-0231-8 Jan Mauer 1, 2 , Miriam Sindelar 1 , Vladimir Despic 1 , Théo Guez 3 , Ben R Hawley 1 , Jean-Jacques Vasseur 3 , Andrea Rentmeister 4 , Steven S Gross 1 , Livio Pellizzoni 5 , Françoise Debart 3 , Hani Goodarzi 6 , Samie R Jaffrey 1
Affiliation
|
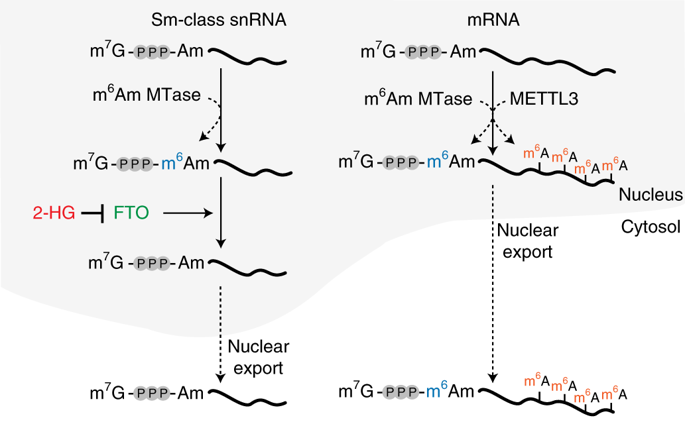
Small nuclear RNAs (snRNAs) are core spliceosome components and mediate pre-mRNA splicing. Here we show that snRNAs contain a regulated and reversible nucleotide modification causing them to exist as two different methyl isoforms, m1 and m2, reflecting the methylation state of the adenosine adjacent to the snRNA cap. We find that snRNA biogenesis involves the formation of an initial m1 isoform with a single-methylated adenosine (2'-O-methyladenosine, Am), which is then converted to a dimethylated m2 isoform (N6,2'-O-dimethyladenosine, m6Am). The relative m1 and m2 isoform levels are determined by the RNA demethylase FTO, which selectively demethylates the m2 isoform. We show FTO is inhibited by the oncometabolite D-2-hydroxyglutarate, resulting in increased m2-snRNA levels. Furthermore, cells that exhibit high m2-snRNA levels show altered patterns of alternative splicing. Together, these data reveal that FTO controls a previously unknown central step of snRNA processing involving reversible methylation, and suggest that epitranscriptomic information in snRNA may influence mRNA splicing.
中文翻译:

FTO在snRNA生物发生过程中控制可逆的m6Am RNA甲基化。
小核RNA(snRNA)是核心剪接体组件,并介导mRNA前剪接。在这里,我们显示snRNA包含受调节且可逆的核苷酸修饰,导致它们以两种不同的甲基亚型m1和m2的形式存在,反映了与snRNA帽相邻的腺苷的甲基化状态。我们发现snRNA生物发生涉及与单个甲基化的腺苷(2'-O-甲基腺苷,Am)的初始m1亚型的形成,然后将其转换为二甲基化的m2同种型(N6,2'-O-二甲基腺苷,m6Am )。相对的m1和m2同工型水平由RNA脱甲基酶FTO确定,该酶选择性地使m2同工型脱甲基。我们显示FTO被oncometabolite D-2-hydroxyglutarate抑制,导致m2-snRNA水平升高。此外,表现出高m2-snRNA水平的细胞显示出可变的剪接模式。总之,这些数据揭示了FTO控制着先前未知的涉及可逆甲基化的snRNA处理的核心步骤,并表明snRNA中的转录组信息可能会影响mRNA的剪接。
更新日期:2019-02-19
中文翻译:

FTO在snRNA生物发生过程中控制可逆的m6Am RNA甲基化。
小核RNA(snRNA)是核心剪接体组件,并介导mRNA前剪接。在这里,我们显示snRNA包含受调节且可逆的核苷酸修饰,导致它们以两种不同的甲基亚型m1和m2的形式存在,反映了与snRNA帽相邻的腺苷的甲基化状态。我们发现snRNA生物发生涉及与单个甲基化的腺苷(2'-O-甲基腺苷,Am)的初始m1亚型的形成,然后将其转换为二甲基化的m2同种型(N6,2'-O-二甲基腺苷,m6Am )。相对的m1和m2同工型水平由RNA脱甲基酶FTO确定,该酶选择性地使m2同工型脱甲基。我们显示FTO被oncometabolite D-2-hydroxyglutarate抑制,导致m2-snRNA水平升高。此外,表现出高m2-snRNA水平的细胞显示出可变的剪接模式。总之,这些数据揭示了FTO控制着先前未知的涉及可逆甲基化的snRNA处理的核心步骤,并表明snRNA中的转录组信息可能会影响mRNA的剪接。

































 京公网安备 11010802027423号
京公网安备 11010802027423号