Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification and characterization of essential genes in the human genome
Science ( IF 44.7 ) Pub Date : 2015-10-15 , DOI: 10.1126/science.aac7041
Tim Wang 1 , Kıvanç Birsoy 1 , Nicholas W Hughes 2 , Kevin M Krupczak 3 , Yorick Post 3 , Jenny J Wei 4 , Eric S Lander 5 , David M Sabatini 6
Science ( IF 44.7 ) Pub Date : 2015-10-15 , DOI: 10.1126/science.aac7041
Tim Wang 1 , Kıvanç Birsoy 1 , Nicholas W Hughes 2 , Kevin M Krupczak 3 , Yorick Post 3 , Jenny J Wei 4 , Eric S Lander 5 , David M Sabatini 6
Affiliation
|
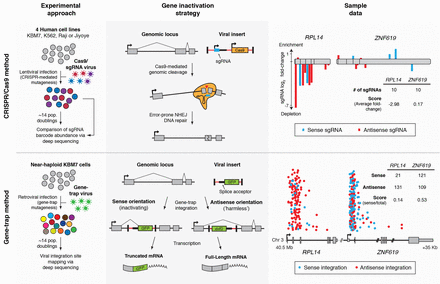
Zeroing in on essential human genes More powerful genetic techniques are helping to define the list of genes required for the life of a human cell. Two papers used the CRISPR genome editing system and a gene trap method in haploid human cells to screen for essential genes (see the Perspective by Boone and Andrews). Wang et al.'s analysis of multiple cell lines indicates that it may be possible to find tumor-specific dependencies on particular genes. Blomen et al. investigate the phenomenon in which nonessential genes are required for fitness in the absence of another gene. Hence, complexity rather than robustness is the human strategy. Science, this issue p. 1096 and p. 1092; see also p. 1028 A comprehensive screen identifies genes required for normal and cancerous human cells to keep growing. [Also see Perspective by Boone and Andrews] Large-scale genetic analysis of lethal phenotypes has elucidated the molecular underpinnings of many biological processes. Using the bacterial clustered regularly interspaced short palindromic repeats (CRISPR) system, we constructed a genome-wide single-guide RNA library to screen for genes required for proliferation and survival in a human cancer cell line. Our screen revealed the set of cell-essential genes, which was validated with an orthogonal gene-trap–based screen and comparison with yeast gene knockouts. This set is enriched for genes that encode components of fundamental pathways, are expressed at high levels, and contain few inactivating polymorphisms in the human population. We also uncovered a large group of uncharacterized genes involved in RNA processing, a number of whose products localize to the nucleolus. Last, screens in additional cell lines showed a high degree of overlap in gene essentiality but also revealed differences specific to each cell line and cancer type that reflect the developmental origin, oncogenic drivers, paralogous gene expression pattern, and chromosomal structure of each line. These results demonstrate the power of CRISPR-based screens and suggest a general strategy for identifying liabilities in cancer cells.
中文翻译:

人类基因组中必需基因的鉴定和表征
瞄准人类必需基因 更强大的遗传技术正在帮助定义人类细胞生命所需的基因列表。两篇论文使用 CRISPR 基因组编辑系统和单倍体人类细胞中的基因捕获方法来筛选必需基因(参见 Boone 和 Andrews 的 Perspective)。 Wang等人对多个细胞系的分析表明,有可能发现肿瘤对特定基因的特异性依赖性。布洛门等人。研究在缺乏其他基因的情况下需要非必需基因来维持健康的现象。因此,人类的策略是复杂性而不是鲁棒性。科学,本期第 14 页。 1096 和 p。 1092;另见 p. 1028 全面筛选确定了正常和癌细胞保持生长所需的基因。 [另请参阅布恩和安德鲁斯的观点]致命表型的大规模遗传分析已经阐明了许多生物过程的分子基础。利用细菌成簇规则间隔短回文重复序列 (CRISPR) 系统,我们构建了全基因组单引导 RNA 文库,以筛选人类癌细胞系增殖和生存所需的基因。我们的筛选揭示了一组细胞必需基因,并通过基于正交基因陷阱的筛选以及与酵母基因敲除的比较进行了验证。该组富含编码基本途径成分的基因,在人群中高水平表达并且包含很少的失活多态性。我们还发现了大量参与 RNA 加工的未表征基因,其中许多产物定位于核仁。 最后,对其他细胞系的筛选显示出基因必要性的高度重叠,但也揭示了每种细胞系和癌症类型特有的差异,这些差异反映了每个细胞系的发育起源、致癌驱动因素、旁系同源基因表达模式和染色体结构。这些结果证明了基于 CRISPR 的筛选的力量,并提出了识别癌细胞中缺陷的一般策略。
更新日期:2015-10-15
中文翻译:

人类基因组中必需基因的鉴定和表征
瞄准人类必需基因 更强大的遗传技术正在帮助定义人类细胞生命所需的基因列表。两篇论文使用 CRISPR 基因组编辑系统和单倍体人类细胞中的基因捕获方法来筛选必需基因(参见 Boone 和 Andrews 的 Perspective)。 Wang等人对多个细胞系的分析表明,有可能发现肿瘤对特定基因的特异性依赖性。布洛门等人。研究在缺乏其他基因的情况下需要非必需基因来维持健康的现象。因此,人类的策略是复杂性而不是鲁棒性。科学,本期第 14 页。 1096 和 p。 1092;另见 p. 1028 全面筛选确定了正常和癌细胞保持生长所需的基因。 [另请参阅布恩和安德鲁斯的观点]致命表型的大规模遗传分析已经阐明了许多生物过程的分子基础。利用细菌成簇规则间隔短回文重复序列 (CRISPR) 系统,我们构建了全基因组单引导 RNA 文库,以筛选人类癌细胞系增殖和生存所需的基因。我们的筛选揭示了一组细胞必需基因,并通过基于正交基因陷阱的筛选以及与酵母基因敲除的比较进行了验证。该组富含编码基本途径成分的基因,在人群中高水平表达并且包含很少的失活多态性。我们还发现了大量参与 RNA 加工的未表征基因,其中许多产物定位于核仁。 最后,对其他细胞系的筛选显示出基因必要性的高度重叠,但也揭示了每种细胞系和癌症类型特有的差异,这些差异反映了每个细胞系的发育起源、致癌驱动因素、旁系同源基因表达模式和染色体结构。这些结果证明了基于 CRISPR 的筛选的力量,并提出了识别癌细胞中缺陷的一般策略。

































 京公网安备 11010802027423号
京公网安备 11010802027423号