npj Genomic Medicine ( IF 4.7 ) Pub Date : 2017-11-01 , DOI: 10.1038/s41525-017-0034-3 Haiyan E Liu 1 , Melanie Triboulet 2 , Amin Zia 3 , Meghah Vuppalapaty 1 , Evelyn Kidess-Sigal 2, 4 , John Coller 5 , Vanita S Natu 5 , Vida Shokoohi 5 , James Che 1 , Corinne Renier 1 , Natalie H Chan 2 , Violet R Hanft 2 , Stefanie S Jeffrey 2 , Elodie Sollier-Christen 1
|
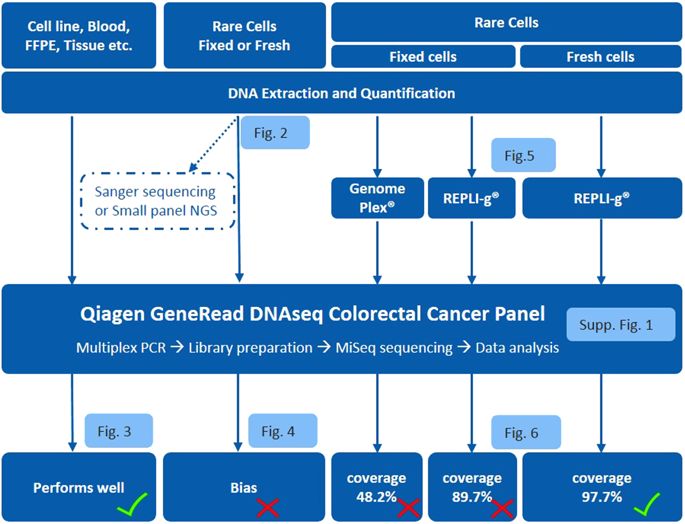
Genomic characterization of circulating tumor cells (CTCs) may prove useful as a surrogate for conventional tissue biopsies. This is particularly important as studies have shown different mutational profiles between CTCs and ctDNA in some tumor subtypes. However, isolating rare CTCs from whole blood has significant hurdles. Very limited DNA quantities often can’t meet NGS requirements without whole genome amplification (WGA). Moreover, white blood cells (WBC) germline contamination may confound CTC somatic mutation analyses. Thus, a good CTC enrichment platform with an efficient WGA and NGS workflow are needed. Here, Vortex label-free CTC enrichment platform was used to capture CTCs. DNA extraction was optimized, WGA evaluated and targeted NGS tested. We used metastatic colorectal cancer (CRC) as the clinical target, HCT116 as the corresponding cell line, GenomePlex® and REPLI-g as the WGA methods, GeneRead DNAseq Human CRC Panel as the 38 gene panel. The workflow was further validated on metastatic CRC patient samples, assaying both tumor and CTCs. WBCs from the same patients were included to eliminate germline contaminations. The described workflow performed well on samples with sufficient DNA, but showed bias for rare cells with limited DNA input. REPLI-g provided an unbiased amplification on fresh rare cells, enabling an accurate variant calling using the targeted NGS. Somatic variants were detected in patient CTCs and not found in age matched healthy donors. This demonstrates the feasibility of a simple workflow for clinically relevant monitoring of tumor genetics in real time and over the course of a patient’s therapy using CTCs.
中文翻译:

用于 CTC 突变检测的全基因组扩增和靶向测序的工作流程优化
循环肿瘤细胞 (CTC) 的基因组特征可能可以作为传统组织活检的替代方法。这一点尤其重要,因为研究表明,在某些肿瘤亚型中,CTC 和 ctDNA 之间存在不同的突变谱。然而,从全血中分离稀有 CTC 存在重大障碍。如果没有全基因组扩增 (WGA),非常有限的 DNA 数量通常无法满足 NGS 要求。此外,白细胞 (WBC) 种系污染可能会混淆 CTC 体细胞突变分析。因此,需要一个具有高效 WGA 和 NGS 工作流程的良好 CTC 富集平台。此处,使用 Vortex 无标记 CTC 富集平台捕获 CTC。优化 DNA 提取、WGA 评估并测试靶向 NGS。我们使用转移性结直肠癌(CRC)作为临床靶点,HCT116作为相应的细胞系,GenomePlex®和REPLI-g作为WGA方法,GeneRead DNAseq Human CRC Panel作为38基因panel。该工作流程在转移性 CRC 患者样本上得到进一步验证,可检测肿瘤和 CTC。来自同一患者的白细胞也被纳入其中,以消除种系污染。所描述的工作流程在具有足够 DNA 的样本上表现良好,但对 DNA 输入有限的稀有细胞表现出偏差。 REPLI-g 对新鲜稀有细胞进行无偏扩增,从而能够使用目标 NGS 进行准确的变异识别。在患者 CTC 中检测到体细胞变异,但在年龄匹配的健康捐赠者中未发现体细胞变异。这证明了在使用 CTC 的患者治疗过程中实时监测肿瘤遗传学临床相关的简单工作流程的可行性。































 京公网安备 11010802027423号
京公网安备 11010802027423号