当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Effect of T·T Mismatch on DNA Dynamics Probed by Minor Groove Binders: Comparison of Dynamic Stokes Shifts of Hoechst and DAPI
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2017-10-02 00:00:00 , DOI: 10.1021/acs.jpcb.7b06937 Him Shweta 1 , Moirangthem Kiran Singh 1 , Kavita Yadav 1 , Sachin Dev Verma 1 , Nibedita Pal 1 , Sobhan Sen 1
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2017-10-02 00:00:00 , DOI: 10.1021/acs.jpcb.7b06937 Him Shweta 1 , Moirangthem Kiran Singh 1 , Kavita Yadav 1 , Sachin Dev Verma 1 , Nibedita Pal 1 , Sobhan Sen 1
Affiliation
|
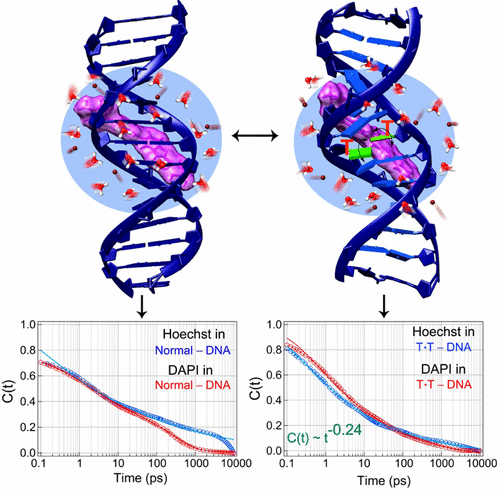
Recognition of DNA base mismatches and their subsequent repair by enzymes is vital for genomic stability. However, it is difficult to comprehend such a process in which enzymes sense and repair different types of mismatches with different ability. It has been suggested that the differential structural changes of mismatched bases act as cues to the repair enzymes, although the effect of such DNA structural changes on surrounding water and ion dynamics is inevitable due to strong electrostatic coupling among them. Thus, collective dynamics of DNA, water, and ions near the mismatch site is believed to be important for mismatch recognition and repair mechanism. Here we show that introduction of a T·T mismatch in the minor groove of DNA induces dispersed (collective) power-law solvation dynamics (of exponent ∼0.24), measured by monitoring the time-resolved fluorescence Stokes shifts (TRFSS) of two popular minor groove binders (Hoechst 33258 and DAPI) over five decades of time from 100 fs to 10 ns. The same ligands however sense different dynamics (power-law of exponent ∼0.15 or power-law multiplied with biexponential relaxation) in the minor groove of normal-DNA. The similar fluorescence anisotropy decays of ligands measured in normal- and T·T-DNA suggest that Stokes shift dynamics and their changes in T·T-DNA purely originate from the solvation process, and not from any internal rotational motion of probe-ligands. The dispersed power-law solvation dynamics seen in T·T-DNA indicate that the ligands do not sense any particular (exponential) relaxation specific to T·T wobbling and/or other conformational changes. This could be the reason why T·T mismatch is recognized by enzymes with lower efficiency compared to purine–pyrimidine and purine–purine mismatches.
中文翻译:

T·T不匹配对小沟结合剂探测DNA动力学的影响:Hoechst和DAPI的动态斯托克斯位移比较
识别DNA碱基错配及其随后被酶修复对于基因组稳定性至关重要。然而,难以理解酶以不同的能力感测和修复不同类型的错配的过程。已经提出错配碱基的差异结构变化是修复酶的线索,尽管由于它们之间强的静电耦合,这种DNA结构变化对周围水和离子动力学的影响是不可避免的。因此,据信错配位点附近的DNA,水和离子的集体动力学对于错配识别和修复机制很重要。在这里,我们表明在DNA的小沟中引入T·T不匹配会诱导分散的(集体)幂律溶剂化动力学(指数约为0.24),通过监测两种流行的小沟结合剂(Hoechst 33258和DAPI)在100 fs至10 ns的五十年时间内的时间分辨荧光斯托克斯位移(TRFSS)进行测量。然而,相同的配体在正常DNA的小沟中感觉到不同的动力学(指数的幂律〜0.15或幂律乘以双指数弛豫)。在正常DNA和T·T-DNA中测得的配体相似的荧光各向异性衰减表明,斯托克斯位移动力学及其在T·T-DNA中的变化完全源自溶剂化过程,而不是源自探针配体的任何内部旋转运动。在T·T-DNA中观察到的分散的幂律溶剂化动力学表明,配体没有感觉到T·T摆动和/或其他构象变化特有的任何特定(指数)弛豫。
更新日期:2017-10-03
中文翻译:

T·T不匹配对小沟结合剂探测DNA动力学的影响:Hoechst和DAPI的动态斯托克斯位移比较
识别DNA碱基错配及其随后被酶修复对于基因组稳定性至关重要。然而,难以理解酶以不同的能力感测和修复不同类型的错配的过程。已经提出错配碱基的差异结构变化是修复酶的线索,尽管由于它们之间强的静电耦合,这种DNA结构变化对周围水和离子动力学的影响是不可避免的。因此,据信错配位点附近的DNA,水和离子的集体动力学对于错配识别和修复机制很重要。在这里,我们表明在DNA的小沟中引入T·T不匹配会诱导分散的(集体)幂律溶剂化动力学(指数约为0.24),通过监测两种流行的小沟结合剂(Hoechst 33258和DAPI)在100 fs至10 ns的五十年时间内的时间分辨荧光斯托克斯位移(TRFSS)进行测量。然而,相同的配体在正常DNA的小沟中感觉到不同的动力学(指数的幂律〜0.15或幂律乘以双指数弛豫)。在正常DNA和T·T-DNA中测得的配体相似的荧光各向异性衰减表明,斯托克斯位移动力学及其在T·T-DNA中的变化完全源自溶剂化过程,而不是源自探针配体的任何内部旋转运动。在T·T-DNA中观察到的分散的幂律溶剂化动力学表明,配体没有感觉到T·T摆动和/或其他构象变化特有的任何特定(指数)弛豫。






























 京公网安备 11010802027423号
京公网安备 11010802027423号