当前位置:
X-MOL 学术
›
ChemistrySelect
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Understanding Chemico‐Biological Interactions of Glutamate MMP‐2 Inhibitors through Rigorous Alignment‐Dependent 3D‐QSAR Analyses
ChemistrySelect ( IF 1.9 ) Pub Date : 2017-09-12 , DOI: 10.1002/slct.201701330 Nilanjan Adhikari 1 , Sk Abdul Amin 1 , Achintya Saha 2 , Tarun Jha 1
ChemistrySelect ( IF 1.9 ) Pub Date : 2017-09-12 , DOI: 10.1002/slct.201701330 Nilanjan Adhikari 1 , Sk Abdul Amin 1 , Achintya Saha 2 , Tarun Jha 1
Affiliation
|
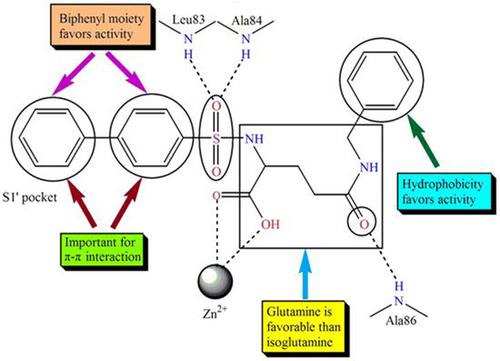
The current study involves different alignment techniques (pharmacophore‐based, docking‐based and Open3DALIGN‐based) extensively at the first time to build three‐dimensional quantitative structure‐activity relationship (3D‐QSAR) models on some potential glutamate‐based in house MMP‐2 inhibitors. The docking‐based alignment method yields the best 3D‐QSAR models though other alignment‐based methods also provide statistically significant and validated models. 3D‐QSAR models are correlated with molecular docking study and pharmacophore mapping. Biphenyl moiety of glutamines enters into the S1′ pocket and involves π‐π interactions with Tyr142 and His120, while none of the phenylacetyl/naphthylacetyl isoglutamines interacts with Tyr142. 3D‐QSAR models also suggest the importance of hydrophobicity and steric features of biphenyl function required for higher MMP‐2 inhibition. The sulfonyl oxygen may form potential hydrogen bonding interaction with Leu83 and Ala84 of S1' pocket. This study clearly points out that biphenylsulfonyl glutamines are favorable than phenylacetyl/naphthylacetyl isoglutamines for imparting higher MMP‐2 inhibition.
中文翻译:

通过严格的比对依赖性3D-QSAR分析了解谷氨酸MMP-2抑制剂的化学-生物相互作用
当前的研究首次广泛涉及不同的比对技术(基于药效基,对接基和基于Open3DALIGN),以在内部一些潜在的基于谷氨酸的基础上建立三维定量结构-活性关系(3D-QSAR)模型MMP-2抑制剂。基于对接的比对方法可产生最佳的3D-QSAR模型,尽管其他基于比对的方法也可提供具有统计意义且经过验证的模型。3D-QSAR模型与分子对接研究和药效团作图相关。谷氨酰胺的联苯部分进入S1'口袋,并与Tyr142和His120进行π-π相互作用,而苯基乙酰基/萘乙酰基异谷氨酰胺均不与Tyr142相互作用。3D-QSAR模型还表明了更高的MMP-2抑制作用所需的疏水性和联苯功能的空间特征的重要性。磺酰氧可与S1'口袋的Leu83和Ala84形成潜在的氢键相互作用。
更新日期:2017-09-12
中文翻译:

通过严格的比对依赖性3D-QSAR分析了解谷氨酸MMP-2抑制剂的化学-生物相互作用
当前的研究首次广泛涉及不同的比对技术(基于药效基,对接基和基于Open3DALIGN),以在内部一些潜在的基于谷氨酸的基础上建立三维定量结构-活性关系(3D-QSAR)模型MMP-2抑制剂。基于对接的比对方法可产生最佳的3D-QSAR模型,尽管其他基于比对的方法也可提供具有统计意义且经过验证的模型。3D-QSAR模型与分子对接研究和药效团作图相关。谷氨酰胺的联苯部分进入S1'口袋,并与Tyr142和His120进行π-π相互作用,而苯基乙酰基/萘乙酰基异谷氨酰胺均不与Tyr142相互作用。3D-QSAR模型还表明了更高的MMP-2抑制作用所需的疏水性和联苯功能的空间特征的重要性。磺酰氧可与S1'口袋的Leu83和Ala84形成潜在的氢键相互作用。

































 京公网安备 11010802027423号
京公网安备 11010802027423号