当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Tryptic Peptides Bearing C-Terminal Dimethyllysine Need to Be Considered during the Analysis of Lysine Dimethylation in Proteomic Study
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-08-08 00:00:00 , DOI: 10.1021/acs.jproteome.7b00373
Ming Chen 1, 2 , Min Zhang 1, 2 , Linhui Zhai 1 , Hao Hu 1 , Ping Liu 1 , Minjia Tan 1, 2
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-08-08 00:00:00 , DOI: 10.1021/acs.jproteome.7b00373
Ming Chen 1, 2 , Min Zhang 1, 2 , Linhui Zhai 1 , Hao Hu 1 , Ping Liu 1 , Minjia Tan 1, 2
Affiliation
|
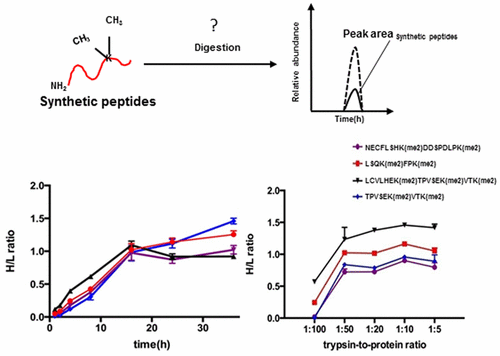
Lysine methylation plays important roles in structural and functional regulation of chromatin. Although trypsin is the most widely used protease in mass spectrometry-based proteomic analysis for lysine methylation substrates, the proteolytic activity of trypsin on dimethylated lysine residues remains an arguable issue. In this study, we tested the ability of trypsin to cleave dimethylated lysine residues in synthetic peptides, purified albumin, and whole cell lysate, and found that the C-terminal of dimethylated lysine residue could be cleaved in a protein sequence-dependent manner. Kinetic studies revealed that the optimal digestion time and enzyme-to-substrate ratio for the cleavage of dimethylated lysine by trypsin was around 16 h and 1:50, respectively. We further showed the tryptic C-terminal lysine-dimethylated (C-Kme2) peptides could contribute to a significant portion of substrate identification in the proteomic study, which utilizes the chemical dimethylation labeling approach. More than 120 tryptic C-Kme2 peptides (7% of total peptides identified) were identified in chemically lysine-dimethyl-labeled HeLa whole cell lysate by a single-shot nanoflow high performance liquid chromatography with tandem mass spectrometry (nano-HPLC–MS/MS) analysis. Moreover, in an assay for substrate identification of protease Glu-C using stable isotope dimethyl labeling approach, our data showed the tryptic C-Kme2 peptides accounted for more than 13% of total tryptic peptides. Additionally, our in vivo methylome profiling data revealed some C-Kme2 peptides, which is of importance to identification and quantification of biologically relevant protein and lysine-methylated site. Therefore, we reason that the tryptic peptides bearing C-terminal dimethylated lysine need to be considered in the mass spectrometric analysis of lysine dimethylation.
中文翻译:

蛋白质组学研究中的赖氨酸二甲基化分析中需要考虑带有C端二甲基赖氨酸的胰蛋白酶肽
赖氨酸甲基化在染色质的结构和功能调节中起重要作用。尽管胰蛋白酶是赖氨酸甲基化底物的基于质谱的蛋白质组学分析中使用最广泛的蛋白酶,但是胰蛋白酶对二甲基化赖氨酸残基的蛋白水解活性仍然是一个有争议的问题。在这项研究中,我们测试了胰蛋白酶裂解合成肽,纯化的白蛋白和全细胞裂解物中的二甲基化赖氨酸残基的能力,发现二甲基化赖氨酸残基的C末端可以以蛋白质序列依赖性的方式裂解。动力学研究表明,胰蛋白酶裂解二甲基化赖氨酸的最佳消化时间和酶底物比分别约为16 h和1:50。我们进一步表明,在蛋白质组学研究中,胰蛋白酶C末端赖氨酸二甲基化(C-Kme2)肽可能有助于底物鉴定的重要部分,该研究利用了化学二甲基化标记方法。通过单次纳米流高效液相色谱-串联质谱法在化学赖氨酸-二甲基标记的HeLa全细胞裂解物中鉴定出120多种胰蛋白酶C-Kme2肽(已鉴定的总肽的7%) MS)分析。此外,在使用稳定同位素二甲基标记方法对蛋白酶Glu-C进行底物鉴定的测定中,我们的数据显示,胰蛋白酶C-Kme2肽占胰蛋白酶总肽的13%以上。此外,我们的体内甲基化组分析数据还显示了一些C-Kme2肽,这对于鉴定和定量生物学相关蛋白质和赖氨酸甲基化位点非常重要。因此,我们认为在赖氨酸二甲基化的质谱分析中需要考虑带有C端二甲基化赖氨酸的胰蛋白酶肽。
更新日期:2017-08-08
中文翻译:

蛋白质组学研究中的赖氨酸二甲基化分析中需要考虑带有C端二甲基赖氨酸的胰蛋白酶肽
赖氨酸甲基化在染色质的结构和功能调节中起重要作用。尽管胰蛋白酶是赖氨酸甲基化底物的基于质谱的蛋白质组学分析中使用最广泛的蛋白酶,但是胰蛋白酶对二甲基化赖氨酸残基的蛋白水解活性仍然是一个有争议的问题。在这项研究中,我们测试了胰蛋白酶裂解合成肽,纯化的白蛋白和全细胞裂解物中的二甲基化赖氨酸残基的能力,发现二甲基化赖氨酸残基的C末端可以以蛋白质序列依赖性的方式裂解。动力学研究表明,胰蛋白酶裂解二甲基化赖氨酸的最佳消化时间和酶底物比分别约为16 h和1:50。我们进一步表明,在蛋白质组学研究中,胰蛋白酶C末端赖氨酸二甲基化(C-Kme2)肽可能有助于底物鉴定的重要部分,该研究利用了化学二甲基化标记方法。通过单次纳米流高效液相色谱-串联质谱法在化学赖氨酸-二甲基标记的HeLa全细胞裂解物中鉴定出120多种胰蛋白酶C-Kme2肽(已鉴定的总肽的7%) MS)分析。此外,在使用稳定同位素二甲基标记方法对蛋白酶Glu-C进行底物鉴定的测定中,我们的数据显示,胰蛋白酶C-Kme2肽占胰蛋白酶总肽的13%以上。此外,我们的体内甲基化组分析数据还显示了一些C-Kme2肽,这对于鉴定和定量生物学相关蛋白质和赖氨酸甲基化位点非常重要。因此,我们认为在赖氨酸二甲基化的质谱分析中需要考虑带有C端二甲基化赖氨酸的胰蛋白酶肽。

































 京公网安备 11010802027423号
京公网安备 11010802027423号