Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
High diversity and unique composition of gut microbiomes in pygmy (Kogia breviceps) and dwarf (K. sima) sperm whales.
Scientific Reports ( IF 3.8 ) Pub Date : 2017-Aug-03 , DOI: 10.1038/s41598-017-07425-z Patrick M. Erwin , Ryan G. Rhodes , Kevin B. Kiser , Tiffany F. Keenan-Bateman , William A. McLellan , D. Ann Pabst
Scientific Reports ( IF 3.8 ) Pub Date : 2017-Aug-03 , DOI: 10.1038/s41598-017-07425-z Patrick M. Erwin , Ryan G. Rhodes , Kevin B. Kiser , Tiffany F. Keenan-Bateman , William A. McLellan , D. Ann Pabst
|
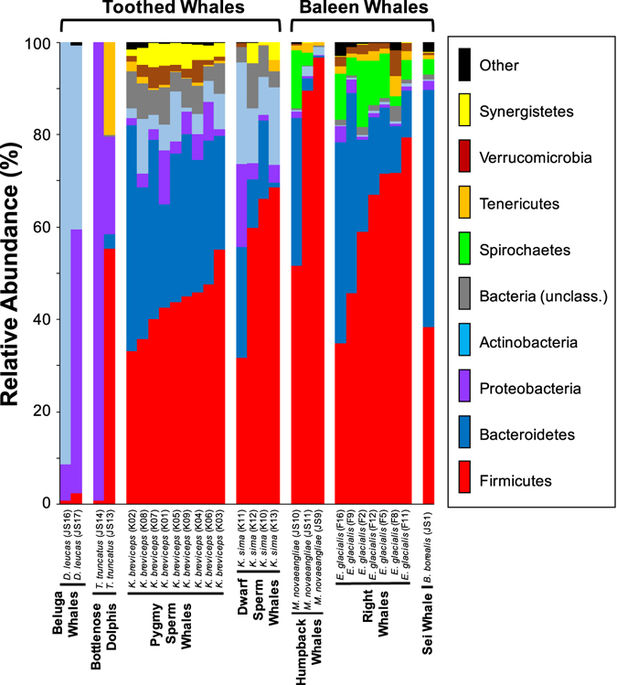
Mammals host diverse bacterial and archaeal symbiont communities (i.e. microbiomes) that play important roles in digestive and immune system functioning, yet cetacean microbiomes remain largely unexplored, in part due to sample collection difficulties. Here, fecal samples from stranded pygmy (Kogia breviceps) and dwarf (K. sima) sperm whales were used to characterize the gut microbiomes of two closely-related species with similar diets. 16S rRNA gene sequencing revealed diverse microbial communities in kogiid whales dominated by Firmicutes and Bacteroidetes. Core symbiont taxa were affiliated with phylogenetic lineages capable of fermentative metabolism and sulfate respiration, indicating potential symbiont contributions to energy acquisition during prey digestion. The diversity and phylum-level composition of kogiid microbiomes differed from those previously reported in toothed whales, which exhibited low diversity communities dominated by Proteobacteria and Actinobacteria. Community structure analyses revealed distinct gut microbiomes in K. breviceps and K. sima, driven by differential relative abundances of shared taxa, and unique microbiomes in kogiid hosts compared to other toothed and baleen whales, driven by differences in symbiont membership. These results provide insight into the diversity, composition and structure of kogiid gut microbiomes and indicate that host identity plays an important role in structuring cetacean microbiomes, even at fine-scale taxonomic levels.
中文翻译:

侏儒抹香鲸(Kogia breviceps)和矮鲸(K. sima)中肠道微生物群的多样性和独特组成。
哺乳动物拥有多种细菌和古细菌共生体群落(即微生物群),它们在消化系统和免疫系统的功能中发挥着重要作用,但是鲸鲨的微生物群在很大程度上仍未得到开发,部分原因是样品采集困难。在这里,来自滞留的侏儒(Kogia breviceps)和侏儒(K. sima)抹香鲸的粪便样本被用来表征两个饮食相似的密切相关物种的肠道微生物群。16S rRNA基因测序揭示了在以纤毛虫和拟杆菌为主导的科吉特鲸中的各种微生物群落。核心共生生物群与能够发酵代谢和硫酸盐呼吸的系统发生谱系相关,表明在猎物消化过程中潜在的共生生物对能量获取的贡献。Kogiid微生物群的多样性和门类组成与以前在齿鲸中报告的那些不同,后者显示出以变形杆菌和放线菌为主导的低多样性群落。群落结构分析显示,与其他带齿鲸和须鲸相比,共生类群的相对相对丰度高,是由K. breviceps和K. sima形成了独特的肠道微生物群,而kogiid宿主中的独特微生物群则是由共生体成员的差异所致。这些结果提供了对kogiid肠道微生物群的多样性,组成和结构的洞察力,并表明宿主身份在构成鲸类微生物群的过程中也起着重要作用,即使是在小规模的生物分类学水平上也是如此。群落结构分析显示,与其他带齿鲸和须鲸相比,共生类群的相对相对丰度高,是由K. breviceps和K. sima产生了独特的肠道微生物群,而在kogiid寄主中,由于共生体成员的差异而导致了独特的微生物群。这些结果提供了对kogiid肠道微生物群的多样性,组成和结构的洞察力,并表明宿主身份在构成鲸类微生物群的过程中也起着重要作用,即使是在小规模的生物分类学水平上也是如此。群落结构分析显示,与其他带齿鲸和须鲸相比,共生类群的相对相对丰度高,是由K. breviceps和K. sima产生了独特的肠道微生物群,而在kogiid寄主中,由于共生体成员的差异而导致了独特的微生物群。这些结果提供了对kogiid肠道微生物群的多样性,组成和结构的洞察力,并表明宿主身份在构成鲸类微生物群的过程中也起着重要作用,即使是在小规模的生物分类学水平上也是如此。
更新日期:2017-08-03
中文翻译:

侏儒抹香鲸(Kogia breviceps)和矮鲸(K. sima)中肠道微生物群的多样性和独特组成。
哺乳动物拥有多种细菌和古细菌共生体群落(即微生物群),它们在消化系统和免疫系统的功能中发挥着重要作用,但是鲸鲨的微生物群在很大程度上仍未得到开发,部分原因是样品采集困难。在这里,来自滞留的侏儒(Kogia breviceps)和侏儒(K. sima)抹香鲸的粪便样本被用来表征两个饮食相似的密切相关物种的肠道微生物群。16S rRNA基因测序揭示了在以纤毛虫和拟杆菌为主导的科吉特鲸中的各种微生物群落。核心共生生物群与能够发酵代谢和硫酸盐呼吸的系统发生谱系相关,表明在猎物消化过程中潜在的共生生物对能量获取的贡献。Kogiid微生物群的多样性和门类组成与以前在齿鲸中报告的那些不同,后者显示出以变形杆菌和放线菌为主导的低多样性群落。群落结构分析显示,与其他带齿鲸和须鲸相比,共生类群的相对相对丰度高,是由K. breviceps和K. sima形成了独特的肠道微生物群,而kogiid宿主中的独特微生物群则是由共生体成员的差异所致。这些结果提供了对kogiid肠道微生物群的多样性,组成和结构的洞察力,并表明宿主身份在构成鲸类微生物群的过程中也起着重要作用,即使是在小规模的生物分类学水平上也是如此。群落结构分析显示,与其他带齿鲸和须鲸相比,共生类群的相对相对丰度高,是由K. breviceps和K. sima产生了独特的肠道微生物群,而在kogiid寄主中,由于共生体成员的差异而导致了独特的微生物群。这些结果提供了对kogiid肠道微生物群的多样性,组成和结构的洞察力,并表明宿主身份在构成鲸类微生物群的过程中也起着重要作用,即使是在小规模的生物分类学水平上也是如此。群落结构分析显示,与其他带齿鲸和须鲸相比,共生类群的相对相对丰度高,是由K. breviceps和K. sima产生了独特的肠道微生物群,而在kogiid寄主中,由于共生体成员的差异而导致了独特的微生物群。这些结果提供了对kogiid肠道微生物群的多样性,组成和结构的洞察力,并表明宿主身份在构成鲸类微生物群的过程中也起着重要作用,即使是在小规模的生物分类学水平上也是如此。















































 京公网安备 11010802027423号
京公网安备 11010802027423号