当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Reconstructing cell cycle pseudo time-series via single-cell transcriptome data.
Nature Communications ( IF 14.7 ) Pub Date : 2017-06-19 , DOI: 10.1038/s41467-017-00039-z
Zehua Liu , Huazhe Lou , Kaikun Xie , Hao Wang , Ning Chen , Oscar M. Aparicio , Michael Q. Zhang , Rui Jiang , Ting Chen
Nature Communications ( IF 14.7 ) Pub Date : 2017-06-19 , DOI: 10.1038/s41467-017-00039-z
Zehua Liu , Huazhe Lou , Kaikun Xie , Hao Wang , Ning Chen , Oscar M. Aparicio , Michael Q. Zhang , Rui Jiang , Ting Chen
|
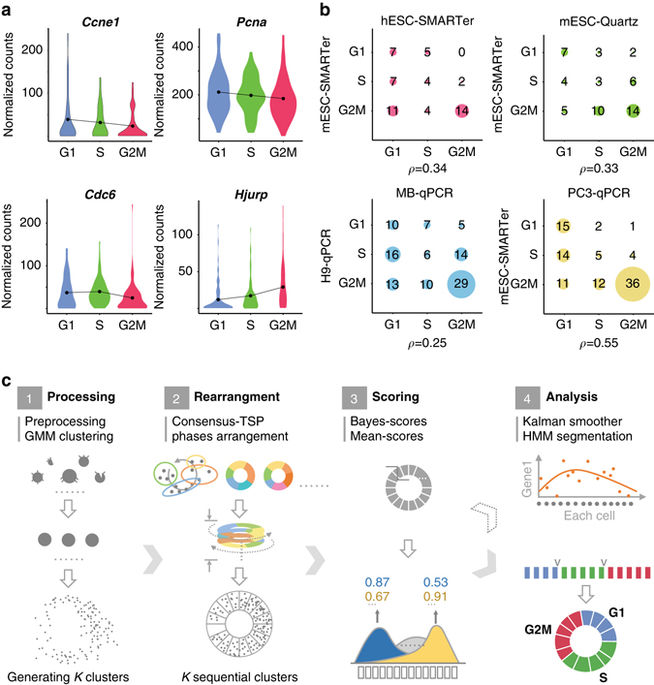
Single-cell mRNA sequencing, which permits whole transcriptional profiling of individual cells, has been widely applied to study growth and development of tissues and tumors. Resolving cell cycle for such groups of cells is significant, but may not be adequately achieved by commonly used approaches. Here we develop a traveling salesman problem and hidden Markov model-based computational method named reCAT, to recover cell cycle along time for unsynchronized single-cell transcriptome data. We independently test reCAT for accuracy and reliability using several data sets. We find that cell cycle genes cluster into two major waves of expression, which correspond to the two well-known checkpoints, G1 and G2. Moreover, we leverage reCAT to exhibit methylation variation along the recovered cell cycle. Thus, reCAT shows the potential to elucidate diverse profiles of cell cycle, as well as other cyclic or circadian processes (e.g., in liver), on single-cell resolution.In single-cell RNA sequencing data of heterogeneous cell populations, cell cycle stage of individual cells would often be informative. Here, the authors introduce a computational model to reconstruct a pseudo-time series from single cell transcriptome data, identify the cell cycle stages, identify candidate cell cycle-regulated genes and recover the methylome changes during the cell cycle.
中文翻译:

通过单细胞转录组数据重建细胞周期伪时间序列。
单细胞mRNA测序允许对单个细胞进行完整的转录分析,已被广泛用于研究组织和肿瘤的生长和发育。解决此类细胞群的细胞周期非常重要,但可能无法通过常用方法充分实现。在这里,我们开发了一个旅行推销员问题和一种基于隐马尔可夫模型的名为reCAT的计算方法,以恢复随时间推移的不同步单细胞转录组数据的细胞周期。我们使用多个数据集独立测试reCAT的准确性和可靠性。我们发现细胞周期基因聚集成两个主要的表达波,分别对应于两个著名的检查点G1和G2。此外,我们利用reCAT在恢复的细胞周期中表现出甲基化变化。因此,reCAT显示了在单细胞分辨率上阐明细胞周期以及其他循环或昼夜节律过程(例如肝脏中)的各种特征的潜力。在异源细胞群体的单细胞RNA测序数据中,个体的细胞周期阶段细胞通常会提供信息。在这里,作者介绍了一种计算模型,可从单细胞转录组数据重建伪时间序列,识别细胞周期阶段,识别候选细胞周期调节基因并恢复细胞周期中甲基化组的变化。
更新日期:2017-06-26
中文翻译:

通过单细胞转录组数据重建细胞周期伪时间序列。
单细胞mRNA测序允许对单个细胞进行完整的转录分析,已被广泛用于研究组织和肿瘤的生长和发育。解决此类细胞群的细胞周期非常重要,但可能无法通过常用方法充分实现。在这里,我们开发了一个旅行推销员问题和一种基于隐马尔可夫模型的名为reCAT的计算方法,以恢复随时间推移的不同步单细胞转录组数据的细胞周期。我们使用多个数据集独立测试reCAT的准确性和可靠性。我们发现细胞周期基因聚集成两个主要的表达波,分别对应于两个著名的检查点G1和G2。此外,我们利用reCAT在恢复的细胞周期中表现出甲基化变化。因此,reCAT显示了在单细胞分辨率上阐明细胞周期以及其他循环或昼夜节律过程(例如肝脏中)的各种特征的潜力。在异源细胞群体的单细胞RNA测序数据中,个体的细胞周期阶段细胞通常会提供信息。在这里,作者介绍了一种计算模型,可从单细胞转录组数据重建伪时间序列,识别细胞周期阶段,识别候选细胞周期调节基因并恢复细胞周期中甲基化组的变化。

































 京公网安备 11010802027423号
京公网安备 11010802027423号