当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Profiling Protease Specificity: Combining Yeast ER Sequestration Screening (YESS) with Next Generation Sequencing
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2017-01-03 00:00:00 , DOI: 10.1021/acschembio.6b00547 Qing Li 1 , Li Yi 1 , Kam Hon Hoi 1 , Peter Marek 1 , George Georgiou 1 , Brent L. Iverson 1
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2017-01-03 00:00:00 , DOI: 10.1021/acschembio.6b00547 Qing Li 1 , Li Yi 1 , Kam Hon Hoi 1 , Peter Marek 1 , George Georgiou 1 , Brent L. Iverson 1
Affiliation
|
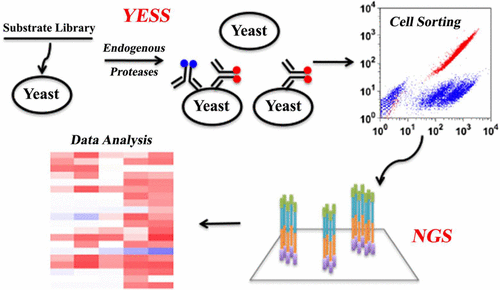
An enzyme engineering technology involving yeast endoplasmic reticulum (ER) sequestration screening (YESS) has been recently developed. Here, a new method is established, in which the YESS platform is combined with NextGen sequencing (NGS) to enable a comprehensive survey of protease specificity. In this approach, a combinatorial substrate library is targeted to the yeast ER and transported through the secretory pathway, interacting with any protease(s) residing in the ER. Multicolor FACS screening is used to isolate cells labeled with fluorophore-conjugated antibodies, followed by NGS to profile the cleaved substrates. The YESS-NGS method was successfully applied to profile the sequence specificity of the wild-type and an engineered variant of the tobacco etch mosaic virus protease. Proteolysis in the yeast secretory pathway was also mapped for the first time in vivo revealing a major cleavage pattern of Ali/Leu-X-Lys/Arg-Arg. Here Ali is any small aliphatic residue, but especially Leu. This pattern was verified to be due to the well-known endogenous protease Kex2 after comparison to a newly generated Kex2 knockout strain as well as cleavage of peptides with recombinant Kex2 in vitro. This information is particularly important for those using yeast display technology, as library members with Ali/Leu-X-Lys/Arg-Arg patterns are likely being removed from screens via Kex2 cleavage without the researcher’s knowledge.
中文翻译:

剖析蛋白酶特异性:结合酵母ER螯合筛选(YESS)与下一代测序
最近开发了一种酶工程技术,涉及酵母内质网(ER)隔离筛选(YESS)。在这里,建立了一种新方法,其中将YESS平台与NextGen测序(NGS)结合使用,以实现蛋白酶特异性的全面调查。在这种方法中,组合底物文库靶向酵母ER,并通过分泌途径转运,与ER中的任何蛋白酶相互作用。多色FACS筛选用于分离标记有荧光团的抗体标记的细胞,然后使用NGS筛选裂解的底物。YESS-NGS方法已成功应用于分析烟草蚀刻花叶病毒蛋白酶的野生型和工程化变异体的序列特异性。体内揭示了Ali / Leu-X-Lys / Arg-Arg的主要切割模式。在这里,Ali是任何小的脂肪族残基,但尤其是Leu。在与新产生的Kex2敲除菌株进行比较后,以及在体外用重组Kex2裂解肽后,证实了这种模式是由于众所周知的内源蛋白酶Kex2 。该信息对于使用酵母展示技术的人们尤其重要,因为具有Ali / Leu-X-Lys / Arg-Arg模式的文库成员可能会在不引起研究人员知识的情况下通过Kex2切割从屏幕上删除。
更新日期:2017-01-03
中文翻译:

剖析蛋白酶特异性:结合酵母ER螯合筛选(YESS)与下一代测序
最近开发了一种酶工程技术,涉及酵母内质网(ER)隔离筛选(YESS)。在这里,建立了一种新方法,其中将YESS平台与NextGen测序(NGS)结合使用,以实现蛋白酶特异性的全面调查。在这种方法中,组合底物文库靶向酵母ER,并通过分泌途径转运,与ER中的任何蛋白酶相互作用。多色FACS筛选用于分离标记有荧光团的抗体标记的细胞,然后使用NGS筛选裂解的底物。YESS-NGS方法已成功应用于分析烟草蚀刻花叶病毒蛋白酶的野生型和工程化变异体的序列特异性。体内揭示了Ali / Leu-X-Lys / Arg-Arg的主要切割模式。在这里,Ali是任何小的脂肪族残基,但尤其是Leu。在与新产生的Kex2敲除菌株进行比较后,以及在体外用重组Kex2裂解肽后,证实了这种模式是由于众所周知的内源蛋白酶Kex2 。该信息对于使用酵母展示技术的人们尤其重要,因为具有Ali / Leu-X-Lys / Arg-Arg模式的文库成员可能会在不引起研究人员知识的情况下通过Kex2切割从屏幕上删除。






























 京公网安备 11010802027423号
京公网安备 11010802027423号