当前位置:
X-MOL 学术
›
Nat. Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition
Nature Chemical Biology ( IF 12.9 ) Pub Date : 2015-03-02 , DOI: 10.1038/nchembio.1768 Christopher D Fage 1 , Eta A Isiorho 1 , Yungnan Liu 2 , Drew T Wagner 1 , Hung-wen Liu 3 , Adrian T Keatinge-Clay 4
中文翻译:

SpnF 的结构,一种催化 [4 + 2] 环加成的独立酶
更新日期:2015-04-04
Nature Chemical Biology ( IF 12.9 ) Pub Date : 2015-03-02 , DOI: 10.1038/nchembio.1768 Christopher D Fage 1 , Eta A Isiorho 1 , Yungnan Liu 2 , Drew T Wagner 1 , Hung-wen Liu 3 , Adrian T Keatinge-Clay 4
Affiliation
|
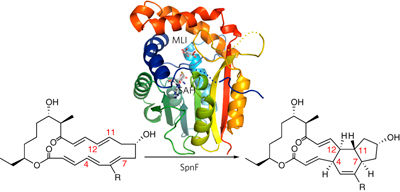
In the biosynthetic pathway of the spinosyn insecticides, the tailoring enzyme SpnF performs a [4 + 2] cycloaddition on a 22-membered macrolactone to forge an embedded cyclohexene ring. To learn more about this reaction, which could potentially proceed through a Diels-Alder mechanism, we determined the 1.50-Å-resolution crystal structure of SpnF bound to S-adenosylhomocysteine. This sets the stage for advanced experimental and computational studies to determine the precise mechanism of SpnF-mediated cyclization.
中文翻译:

SpnF 的结构,一种催化 [4 + 2] 环加成的独立酶
在多杀菌素杀虫剂的生物合成途径中,剪裁酶 SpnF 在 22 元大环内酯上进行 [4 + 2] 环加成,形成嵌入的环己烯环。为了更多地了解这一可能通过 Diels-Alder 机制进行的反应,我们确定了与S-腺苷高半胱氨酸结合的 SpnF 的 1.50 Å 分辨率晶体结构。这为先进的实验和计算研究奠定了基础,以确定 SpnF 介导的环化的精确机制。































 京公网安备 11010802027423号
京公网安备 11010802027423号