当前位置:
X-MOL 学术
›
ACS Med. Chem. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Quantum Mechanics-Based Ranking of Predicted Proteolysis Targeting Chimeras-Mediated Ternary Complexes
ACS Medicinal Chemistry Letters ( IF 3.5 ) Pub Date : 2025-02-06 , DOI: 10.1021/acsmedchemlett.4c00534
Stefania Monteleone 1 , Inaki Morao 2 , Dmitri G Fedorov 3 , Tahsin F Kellici 1
ACS Medicinal Chemistry Letters ( IF 3.5 ) Pub Date : 2025-02-06 , DOI: 10.1021/acsmedchemlett.4c00534
Stefania Monteleone 1 , Inaki Morao 2 , Dmitri G Fedorov 3 , Tahsin F Kellici 1
Affiliation
|
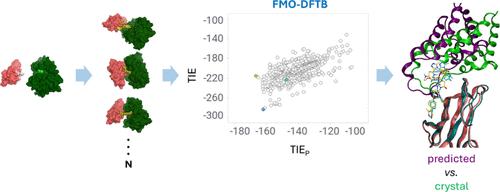
Targeted protein degradation has become the most pursued alternative modality to small-molecule inhibition over the past decade. The traditional strategy of blocking protein activity by tightly binding to a functional substrate pocket has progressed toward proteolysis-targeting chimeras (PROTACs), bivalent molecules that induce the knockdown of targeted proteins. Herein, a combined protocol is described for modeling ternary complexes via well-established approaches. We performed local protein–protein docking using Rosetta protocol and sampled the conformational landscape of a specific PROTAC molecule that was compatible with the generated protein–protein docking poses, followed by double and independent single-linkage/nearest-neighbor clustering for representative selection. Subsequently, we combined the fragment molecular orbital and density functional tight-binding methods to facilitate fast quantum mechanics-based energy calculations of the clustered ternary complexes. Finally, the computed energy values were utilized to score and select the best ternary poses, achieving good agreement with available crystallographic data.
中文翻译:

基于量子力学的针对嵌合体介导的三元复合物的预测蛋白水解排名
靶向蛋白质降解已成为过去十年中小分子抑制最受追捧的替代方式。通过与功能性底物口袋紧密结合来阻断蛋白质活性的传统策略已经发展到蛋白水解靶向嵌合体 (PROTAC),这是一种诱导靶向蛋白质敲低的二价分子。在此,描述了一种通过成熟方法对三元复合物进行建模的组合协议。我们使用 Rosetta 协议进行局部蛋白质-蛋白质对接,并对与生成的蛋白质-蛋白质对接姿势相容的特定 PROTAC 分子的构象景观进行采样,然后进行双重和独立的单联/最近邻聚类以进行代表性选择。随后,我们将片段分子轨道和密度泛函紧密结合方法相结合,以促进基于量子力学的快速能量计算簇状三元复合物。最后,利用计算出的能量值对最佳三元位姿进行评分和选择,与可用的晶体学数据保持一致。
更新日期:2025-02-06
中文翻译:

基于量子力学的针对嵌合体介导的三元复合物的预测蛋白水解排名
靶向蛋白质降解已成为过去十年中小分子抑制最受追捧的替代方式。通过与功能性底物口袋紧密结合来阻断蛋白质活性的传统策略已经发展到蛋白水解靶向嵌合体 (PROTAC),这是一种诱导靶向蛋白质敲低的二价分子。在此,描述了一种通过成熟方法对三元复合物进行建模的组合协议。我们使用 Rosetta 协议进行局部蛋白质-蛋白质对接,并对与生成的蛋白质-蛋白质对接姿势相容的特定 PROTAC 分子的构象景观进行采样,然后进行双重和独立的单联/最近邻聚类以进行代表性选择。随后,我们将片段分子轨道和密度泛函紧密结合方法相结合,以促进基于量子力学的快速能量计算簇状三元复合物。最后,利用计算出的能量值对最佳三元位姿进行评分和选择,与可用的晶体学数据保持一致。

































 京公网安备 11010802027423号
京公网安备 11010802027423号