当前位置:
X-MOL 学术
›
Water Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Enhanced detection for antibiotic resistance genes in wastewater samples using a CRISPR-enriched metagenomic method
Water Research ( IF 11.4 ) Pub Date : 2024-12-26 , DOI: 10.1016/j.watres.2024.123056 Yuqing Mao, Joanna L. Shisler, Thanh H. Nguyen
Water Research ( IF 11.4 ) Pub Date : 2024-12-26 , DOI: 10.1016/j.watres.2024.123056 Yuqing Mao, Joanna L. Shisler, Thanh H. Nguyen
|
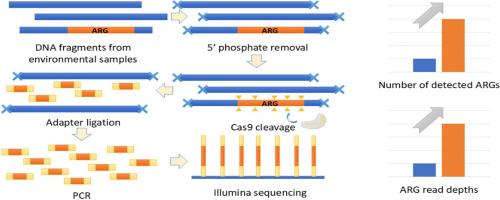
The spread of antibiotic resistance genes (ARGs) in the environment is a global public health concern. To date, over 5,000 genes have been identified to express resistance to antibiotics. ARGs are usually low in abundance for wastewater samples, making them difficult to detect. Metagenomic sequencing and quantitative polymerase chain reaction (qPCR), two conventional ARG detection methods, have low sensitivity and low throughput limitations, respectively. We developed a CRISPR-Cas9-modified next-generation sequencing (NGS) method to enrich the targeted ARGs during library preparation. The false negative and false positive of this method were determined based on a mixture of bacterial isolates with known whole-genome sequences. Low values of both false negative (2/1,208) and false positive (1/1,208) proved the method's reliability. We compared the results obtained by this CRISPR-NGS and the conventional NGS method for six untreated wastewater samples. As compared to the ARGs detected in the same samples using the regular NGS method, the CRISPR-NGS method found up to 1,189 more ARGs and up to 61 more ARG families in low abundances, including the clinically important KPC beta-lactamase genes in the six wastewater samples collected from different sources. Compared to the regular NGS method, the CRISPR-NGS method lowered the detection limit of ARGs from the magnitude of 10-4 to 10-5 as quantified by qPCR relative abundance. The CRISPR-NGS method is promising for ARG detection in wastewater. A similar workflow can also be applied to detect other targets that are in low abundance but of high diversity.
中文翻译:

使用富含 CRISPR 的宏基因组方法增强对废水样品中抗生素耐药基因的检测
抗生素耐药基因 (ARG) 在环境中的传播是一个全球性的公共卫生问题。迄今为止,已鉴定出 5,000 多个表达对抗生素耐药性的基因。废水样品的 ARG 含量通常较低,因此难以检测。宏基因组测序和定量聚合酶链反应 (qPCR) 是两种传统的 ARG 检测方法,分别具有低灵敏度和低通量限制。我们开发了一种 CRISPR-Cas9 修饰的下一代测序 (NGS) 方法,用于在文库制备过程中富集靶向 ARG。该方法的假阴性和假阳性是根据具有已知全基因组序列的细菌分离物的混合物确定的。假阴性 (2/1,208) 和假阳性 (1/1,208) 的低值证明了该方法的可靠性。我们比较了 6 个未经处理的废水样品的 CRISPR-NGS 和常规 NGS 方法获得的结果。与使用常规 NGS 方法在相同样本中检测到的 ARG 相比,CRISPR-NGS 方法发现了多达 1,189 个 ARG 和多达 61 个低丰度的 ARG 家族,包括从不同来源收集的 6 个废水样本中具有临床重要性的 KPC β-内酰胺酶基因。与常规 NGS 方法相比,CRISPR-NGS 方法将 ARGs 的检测限从 10-4 的数量级降低到 10-5,通过 qPCR 相对丰度进行定量。CRISPR-NGS 方法有望用于废水中的 ARG 检测。类似的工作流程也可用于检测丰度低但多样性高的其他靶标。
更新日期:2024-12-27
中文翻译:

使用富含 CRISPR 的宏基因组方法增强对废水样品中抗生素耐药基因的检测
抗生素耐药基因 (ARG) 在环境中的传播是一个全球性的公共卫生问题。迄今为止,已鉴定出 5,000 多个表达对抗生素耐药性的基因。废水样品的 ARG 含量通常较低,因此难以检测。宏基因组测序和定量聚合酶链反应 (qPCR) 是两种传统的 ARG 检测方法,分别具有低灵敏度和低通量限制。我们开发了一种 CRISPR-Cas9 修饰的下一代测序 (NGS) 方法,用于在文库制备过程中富集靶向 ARG。该方法的假阴性和假阳性是根据具有已知全基因组序列的细菌分离物的混合物确定的。假阴性 (2/1,208) 和假阳性 (1/1,208) 的低值证明了该方法的可靠性。我们比较了 6 个未经处理的废水样品的 CRISPR-NGS 和常规 NGS 方法获得的结果。与使用常规 NGS 方法在相同样本中检测到的 ARG 相比,CRISPR-NGS 方法发现了多达 1,189 个 ARG 和多达 61 个低丰度的 ARG 家族,包括从不同来源收集的 6 个废水样本中具有临床重要性的 KPC β-内酰胺酶基因。与常规 NGS 方法相比,CRISPR-NGS 方法将 ARGs 的检测限从 10-4 的数量级降低到 10-5,通过 qPCR 相对丰度进行定量。CRISPR-NGS 方法有望用于废水中的 ARG 检测。类似的工作流程也可用于检测丰度低但多样性高的其他靶标。































 京公网安备 11010802027423号
京公网安备 11010802027423号