当前位置:
X-MOL 学术
›
Adv. Funct. Mater.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Physically‐Based Machine Learning Approach Inspires an Analytical Model for Spider Silk Supercontraction
Advanced Functional Materials ( IF 18.5 ) Pub Date : 2024-12-26 , DOI: 10.1002/adfm.202420095 Vincenzo Fazio, Ali D. Malay, Keiji Numata, Nicola M. Pugno, Giuseppe Puglisi
Advanced Functional Materials ( IF 18.5 ) Pub Date : 2024-12-26 , DOI: 10.1002/adfm.202420095 Vincenzo Fazio, Ali D. Malay, Keiji Numata, Nicola M. Pugno, Giuseppe Puglisi
|
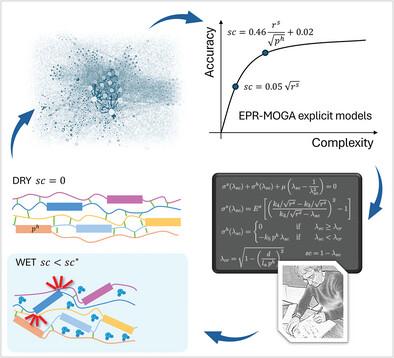
Scientific and industrial interest in spider silk stems from its remarkable properties, including supercontraction—an activation effect induced by wetting. Understanding the underlying molecular scale mechanisms is then also crucial for biomimetic applications. In this study, it is illustrated how the effective integration of physically‐based machine learning with scientific interpretations can lead to significant physical insights and enhance the predictive power of an existing microstructure‐inspired model. A symbolic data modeling technique, known as Evolutionary Polynomial Regression (EPR), is employed, which integrates regression capabilities with the genetic programming paradigm, enabling the derivation of explicit analytical formulas for deducing structure‐function relationships emerging across different scales, to investigate the impact of protein primary structures on supercontraction. This analysis is based on recent multiscale experimental data encompassing a diverse range of scales and a wide variety of different spider silks. Specifically, this analysis reveals a correlation between supercontraction and the repeat length of the MaSp2 protein as well as the polyalanine region of MaSp1. Straightforward microstructural interpretations that align with experimental observations are proposed. The MaSp2 repeat length governs the cross‐links that stabilize amorphous chains in dry conditions. When hydrated, these cross‐links are disrupted, leading to entropic coiling and fiber contraction. Furthermore, the length of the polyalanine region in MaSp1 plays a critical role in supercontraction by restricting the extent of crystal misalignment necessary to accommodate the shortening of the soft regions. The validation of the model is accomplished by comparing experimental data from the Silkome database with theoretical predictions derived from both the machine learning and the proposed model. The enhanced model offers a more comprehensive understanding of supercontraction and establishes a link between the primary structure of silk proteins and their macroscopic behavior, thereby advancing the field of biomimetic applications.
中文翻译:

基于物理的机器学习方法激发了蜘蛛丝超收缩的分析模型
科学界和工业界对蜘蛛丝的兴趣源于其卓越的特性,包括超收缩——一种由润湿引起的活化效应。因此,了解潜在的分子尺度机制对于仿生应用也至关重要。在这项研究中,说明了基于物理的机器学习与科学解释的有效整合如何带来重要的物理见解并增强现有微结构启发模型的预测能力。采用了一种称为进化多项式回归 (EPR) 的符号数据建模技术,该技术将回归功能与遗传编程范式相结合,能够推导出明确的分析公式,用于推断在不同尺度上出现的结构-功能关系,以研究蛋白质一级结构对超收缩的影响。该分析基于最近的多尺度实验数据,包括各种尺度和各种不同的蜘蛛丝。具体来说,该分析揭示了超收缩与 MaSp2 蛋白的重复长度以及 MaSp1 的聚丙氨酸区域之间的相关性。提出了与实验观察结果一致的简单微观结构解释。MaSp2 重复长度控制在干燥条件下稳定无定形链的交联。当水合时,这些交联被破坏,导致熵卷曲和纤维收缩。此外,MaSp1 中聚丙氨酸区域的长度在超收缩中起着关键作用,它限制了适应软区域缩短所需的晶体错位程度。 模型的验证是通过将 Silkome 数据库的实验数据与机器学习和拟议模型得出的理论预测进行比较来完成的。增强的模型提供了对超收缩的更全面理解,并在丝蛋白的一级结构与其宏观行为之间建立了联系,从而推动了仿生应用领域。
更新日期:2024-12-26
中文翻译:

基于物理的机器学习方法激发了蜘蛛丝超收缩的分析模型
科学界和工业界对蜘蛛丝的兴趣源于其卓越的特性,包括超收缩——一种由润湿引起的活化效应。因此,了解潜在的分子尺度机制对于仿生应用也至关重要。在这项研究中,说明了基于物理的机器学习与科学解释的有效整合如何带来重要的物理见解并增强现有微结构启发模型的预测能力。采用了一种称为进化多项式回归 (EPR) 的符号数据建模技术,该技术将回归功能与遗传编程范式相结合,能够推导出明确的分析公式,用于推断在不同尺度上出现的结构-功能关系,以研究蛋白质一级结构对超收缩的影响。该分析基于最近的多尺度实验数据,包括各种尺度和各种不同的蜘蛛丝。具体来说,该分析揭示了超收缩与 MaSp2 蛋白的重复长度以及 MaSp1 的聚丙氨酸区域之间的相关性。提出了与实验观察结果一致的简单微观结构解释。MaSp2 重复长度控制在干燥条件下稳定无定形链的交联。当水合时,这些交联被破坏,导致熵卷曲和纤维收缩。此外,MaSp1 中聚丙氨酸区域的长度在超收缩中起着关键作用,它限制了适应软区域缩短所需的晶体错位程度。 模型的验证是通过将 Silkome 数据库的实验数据与机器学习和拟议模型得出的理论预测进行比较来完成的。增强的模型提供了对超收缩的更全面理解,并在丝蛋白的一级结构与其宏观行为之间建立了联系,从而推动了仿生应用领域。































 京公网安备 11010802027423号
京公网安备 11010802027423号