当前位置:
X-MOL 学术
›
Water Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Deciphering the N1-substituent effects on biodegradation of sulfonamides: Novel insights revealed from molecular biology and computational chemistry approaches
Water Research ( IF 11.4 ) Pub Date : 2024-12-25 , DOI: 10.1016/j.watres.2024.123037 Jiahui Hu, Ruiyang Li, Jiayu Zhang, Lijia Cao, Huaxin Lei, Renxin Zhao, Lin Lin, Xiao-yan Li, Wen Zhang, Bing Li
Water Research ( IF 11.4 ) Pub Date : 2024-12-25 , DOI: 10.1016/j.watres.2024.123037 Jiahui Hu, Ruiyang Li, Jiayu Zhang, Lijia Cao, Huaxin Lei, Renxin Zhao, Lin Lin, Xiao-yan Li, Wen Zhang, Bing Li
|
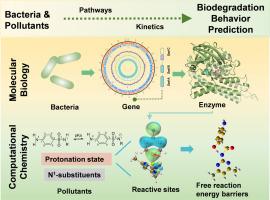
Elucidating biodegradation mechanisms and predicting pollutant reactivities are essential for advancing the application of biodegradation engineering to address the challenge of thousands of emerging contaminants. Molecular biology and computational chemistry are powerful tools for this purpose, enabling the investigation of biochemical reactions at both the gene and atomic levels. This study employs the biodegradation of ten sulfonamide antibiotics as a case study to demonstrate the integration of genomics and quantum chemistry approaches in exploring the biodegradation behavior of emerging contaminants. The isolated functional strain, Paenarthrobacter sp., could completely degrade all ten model sulfonamides under aerobic conditions. These compounds share a 4-aminobenzenesulfonamide core but differ in N1-substituent rings. Despite structural variations, all sulfonamides follow a consistent degradation pathway, yielding aminated heterocycles as end products. This pathway involves key steps such as dehydrogenation activation, ipso-hydroxylation, and the cleavage of S-N and S-C bonds, with the latter being particularly influenced by the N1-substituents. Heterocyclic structures affect biodegradation rates by altering the electronic density at the C3 and N1 atoms of sulfonamides. Substituents with higher electron-donating potential and lower Gibbs free energy barriers for S-C and C-N bond cleavage significantly enhance biodegradation efficiency. This work not only deciphers the universal biodegradation mechanism of sulfonamides but also offers theoretical insights for predicting the biodegradation behavior and pattern of emerging contaminants. These findings contribute to the effective removal of emerging contaminants from aquatic environments, advancing the practical application of biotreatment technologies.
中文翻译:

破译 N1 取代基对磺胺类药物生物降解的影响:从分子生物学和计算化学方法揭示的新见解
阐明生物降解机制和预测污染物反应性对于推进生物降解工程的应用以应对数千种新兴污染物的挑战至关重要。分子生物学和计算化学是实现此目的的强大工具,能够在基因和原子水平上研究生化反应。本研究以 10 种磺胺类抗生素的生物降解为案例研究,展示了基因组学和量子化学方法在探索新兴污染物的生物降解行为方面的整合。分离的功能菌株 Paenarthrobacter sp. 在有氧条件下可以完全降解所有 10 种模型磺胺类药物。这些化合物共享一个 4-氨基苯磺酰胺核心,但在 N1-取代基环上有所不同。尽管结构存在差异,但所有磺胺类药物都遵循一致的降解途径,产生胺化杂环作为最终产物。该途径涉及关键步骤,例如脱氢激活、ipso-羟基化以及 S-N 和 S-C 键的裂解,后者特别受 N1-取代基的影响。杂环结构通过改变磺胺类药物 C3 和 N1 原子的电子密度来影响生物降解速率。具有较高供电子电位和较低 S-C 和 C-N 键裂解的吉布斯自由能垒的取代基可显著提高生物降解效率。这项工作不仅破译了磺胺类药物的普遍生物降解机制,还为预测新兴污染物的生物降解行为和模式提供了理论见解。 这些发现有助于有效去除水生环境中新出现的污染物,推动生物处理技术的实际应用。
更新日期:2024-12-30
中文翻译:

破译 N1 取代基对磺胺类药物生物降解的影响:从分子生物学和计算化学方法揭示的新见解
阐明生物降解机制和预测污染物反应性对于推进生物降解工程的应用以应对数千种新兴污染物的挑战至关重要。分子生物学和计算化学是实现此目的的强大工具,能够在基因和原子水平上研究生化反应。本研究以 10 种磺胺类抗生素的生物降解为案例研究,展示了基因组学和量子化学方法在探索新兴污染物的生物降解行为方面的整合。分离的功能菌株 Paenarthrobacter sp. 在有氧条件下可以完全降解所有 10 种模型磺胺类药物。这些化合物共享一个 4-氨基苯磺酰胺核心,但在 N1-取代基环上有所不同。尽管结构存在差异,但所有磺胺类药物都遵循一致的降解途径,产生胺化杂环作为最终产物。该途径涉及关键步骤,例如脱氢激活、ipso-羟基化以及 S-N 和 S-C 键的裂解,后者特别受 N1-取代基的影响。杂环结构通过改变磺胺类药物 C3 和 N1 原子的电子密度来影响生物降解速率。具有较高供电子电位和较低 S-C 和 C-N 键裂解的吉布斯自由能垒的取代基可显著提高生物降解效率。这项工作不仅破译了磺胺类药物的普遍生物降解机制,还为预测新兴污染物的生物降解行为和模式提供了理论见解。 这些发现有助于有效去除水生环境中新出现的污染物,推动生物处理技术的实际应用。































 京公网安备 11010802027423号
京公网安备 11010802027423号