当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Additive CHARMM Force Field for Pterins and Folates
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-12-23 , DOI: 10.1002/jcc.27548 Elsa Balduzzi, Wenlu Yin, Jean‐Christophe Lambry, Hannu Myllykallio, Alexey Aleksandrov
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-12-23 , DOI: 10.1002/jcc.27548 Elsa Balduzzi, Wenlu Yin, Jean‐Christophe Lambry, Hannu Myllykallio, Alexey Aleksandrov
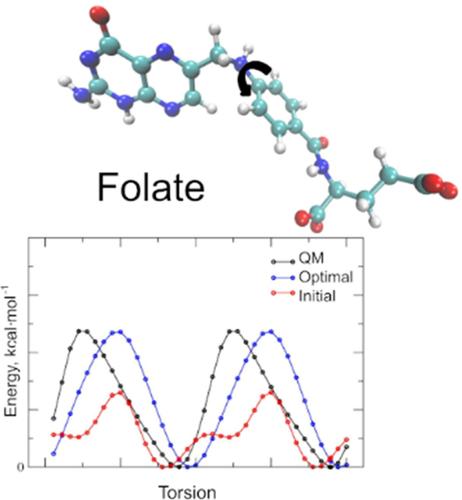
|
Folates comprise a crucial class of biologically active compounds related to folic acid, playing a vital role in numerous enzymatic reactions. One‐carbon metabolism, facilitated by the folate cofactor, supports numerous physiological processes, including biosynthesis, amino acid homeostasis, epigenetic maintenance, and redox defense. Folates share a common pterin heterocyclic ring structure capable of undergoing redox reactions and existing in various protonation states. This study aimed to derive molecular mechanics (MM) parameters compatible with the CHARMM36 all‐atom additive force field for pterins and biologically important folates, including pterin, biopterin, and folic acid. Three redox forms were considered: oxidized, dihydrofolate, and tetrahydrofolate states. Across all protonation states, a total of 18 folates were parameterized. Partial charges were derived using the CHARMM force field parametrization protocol, based on targeting reference quantum mechanics monohydrate interactions, electrostatic potential, and dipole moment. Bonded terms were parameterized using one‐dimensional adiabatic potential energy surface scans, and two‐dimensional scans to parametrize in‐ring torsions associated with the puckering states of dihydropterin and tetrahydropterin. The quality of the model was demonstrated through simulations of three protein complexes using optimized and initial parameters. These simulations underscored the significantly enhanced performance of the folate model developed in this study compared to the initial model without optimization in reproducing structural properties of folate–protein complexes. Overall, the presented MM model will be valuable for modeling folates in various redox states and serve as a starting point for parameterizing other folate derivatives.
中文翻译:

Pterins 和 Folates 的加性 CHARMM 力场
叶酸是一类与叶酸相关的重要生物活性化合物,在许多酶促反应中起着至关重要的作用。由叶酸辅因子促进的一碳代谢支持许多生理过程,包括生物合成、氨基酸稳态、表观遗传维持和氧化还原防御。叶酸盐具有共同的蝶呤杂环结构,能够进行氧化还原反应并以各种质子化状态存在。本研究旨在推导出分子力学 (MM) 参数,该参数与蝶呤和生物学上重要的叶酸(包括蝶呤、生物蝶呤和叶酸)的CHARMM36全原子加性力场兼容。考虑了三种氧化还原形式:氧化、二氢叶酸和四氢叶酸状态。在所有质子化状态中,共有 18 个叶酸被参数化。部分电荷是使用 CHARMM 力场参数化协议得出的,该协议基于参考量子力学一水合物相互作用、静电势和偶极矩。使用一维绝热势能表面扫描对键合项进行参数化,并使用二维扫描来参数化与二氢蝶呤和四氢蝶呤的皱褶状态相关的环内扭转。通过使用优化参数和初始参数对三种蛋白质复合物进行模拟,证明了模型的质量。这些模拟强调了与未优化叶酸-蛋白质复合物结构特性的初始模型相比,本研究中开发的叶酸模型的性能显着增强。 总体而言,所提出的 MM 模型对于模拟各种氧化还原状态下的叶酸很有价值,并作为参数化其他叶酸衍生物的起点。
更新日期:2024-12-23
中文翻译:

Pterins 和 Folates 的加性 CHARMM 力场
叶酸是一类与叶酸相关的重要生物活性化合物,在许多酶促反应中起着至关重要的作用。由叶酸辅因子促进的一碳代谢支持许多生理过程,包括生物合成、氨基酸稳态、表观遗传维持和氧化还原防御。叶酸盐具有共同的蝶呤杂环结构,能够进行氧化还原反应并以各种质子化状态存在。本研究旨在推导出分子力学 (MM) 参数,该参数与蝶呤和生物学上重要的叶酸(包括蝶呤、生物蝶呤和叶酸)的CHARMM36全原子加性力场兼容。考虑了三种氧化还原形式:氧化、二氢叶酸和四氢叶酸状态。在所有质子化状态中,共有 18 个叶酸被参数化。部分电荷是使用 CHARMM 力场参数化协议得出的,该协议基于参考量子力学一水合物相互作用、静电势和偶极矩。使用一维绝热势能表面扫描对键合项进行参数化,并使用二维扫描来参数化与二氢蝶呤和四氢蝶呤的皱褶状态相关的环内扭转。通过使用优化参数和初始参数对三种蛋白质复合物进行模拟,证明了模型的质量。这些模拟强调了与未优化叶酸-蛋白质复合物结构特性的初始模型相比,本研究中开发的叶酸模型的性能显着增强。 总体而言,所提出的 MM 模型对于模拟各种氧化还原状态下的叶酸很有价值,并作为参数化其他叶酸衍生物的起点。































 京公网安备 11010802027423号
京公网安备 11010802027423号