Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mutability and hypermutation antagonize immunoglobulin codon optimality
Molecular Cell ( IF 14.5 ) Pub Date : 2024-12-20 , DOI: 10.1016/j.molcel.2024.11.033 Joshua J.C. McGrath, Juyeon Park, Chloe A. Troxell, Jordan C. Chervin, Lei Li, Johnathan R. Kent, Siriruk Changrob, Yanbin Fu, Min Huang, Nai-Ying Zheng, G. Dewey Wilbanks, Sean A. Nelson, Jiayi Sun, Giorgio Inghirami, Maria Lucia L. Madariaga, George Georgiou, Patrick C. Wilson
Molecular Cell ( IF 14.5 ) Pub Date : 2024-12-20 , DOI: 10.1016/j.molcel.2024.11.033 Joshua J.C. McGrath, Juyeon Park, Chloe A. Troxell, Jordan C. Chervin, Lei Li, Johnathan R. Kent, Siriruk Changrob, Yanbin Fu, Min Huang, Nai-Ying Zheng, G. Dewey Wilbanks, Sean A. Nelson, Jiayi Sun, Giorgio Inghirami, Maria Lucia L. Madariaga, George Georgiou, Patrick C. Wilson
|
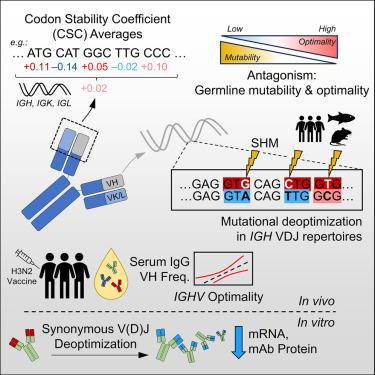
The efficacy of antibody responses is inherently linked to paratope diversity, as generated through V(D)J recombination and somatic hypermutation. Despite this, it is unclear how genetic diversification mechanisms evolved alongside codon optimality and affect antibody expression. Here, we analyze germline immunoglobulin (IG) genes, natural V(D)J repertoires, serum IgG, and monoclonal antibody (mAb) expression through the lens of codon optimality. Germline variable genes (IGVs) exhibit diverse optimality that is inversely related to mutability. Hypermutation deoptimizes heavy-chain (IGH) VDJ repertoires within human tonsils, bone marrow, lymph nodes (including SARS-CoV-2-specific clones), blood (HIV-1-specific clones), mice, and zebrafish. Analyses of mutation-affected codons show that targeting to complementarity-determining regions constrains deoptimization. Germline IGHV optimality correlates with serum variable fragment (VH) usage after influenza vaccination, while synonymous deoptimization attenuated mAb yield. These findings provide unanticipated insights into an antagonistic relationship between diversification mechanisms and codon optimality. Ultimately, the need for diversity takes precedence over that for the most optimal codon usage.
更新日期:2024-12-20






























 京公网安备 11010802027423号
京公网安备 11010802027423号