当前位置:
X-MOL 学术
›
Biotechnol. Bioeng.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Efficient Multiplex Genome Editing of the Cyanobacterium Synechocystis sp. PCC6803 via CRISPR‐Cas12a
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2024-12-20 , DOI: 10.1002/bit.28910 Wei Du, Luna L. Meister, Tobias van Grinsven, Filipe Branco dos Santos
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2024-12-20 , DOI: 10.1002/bit.28910 Wei Du, Luna L. Meister, Tobias van Grinsven, Filipe Branco dos Santos
|
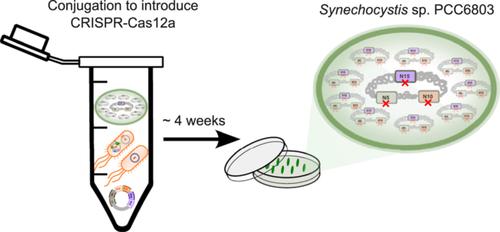
Cyanobacteria have been genetically modified to convert CO2 into biochemical products, but efficient genetic engineering tools, including CRISPR‐Cas systems, remain limited. This is primarily due to the polyploid nature of cyanobacteria, which hinders their effectiveness. Here, we address the latter by specifically (i) modifying the RSF1010‐based replicative plasmid to simplify cloning efforts while maintaining high conjugation efficiency; (ii) improving the design of the guide RNA (gRNA) to facilitate chromosomal cleavage; (iii) introducing template DNA fragments as pure plasmids via natural transformation; and (iv) using sacB to facilitate replicative plasmid curing. With this system, the replicative plasmid containing both Cas12a and gRNA is introduced to Synechocystis sp. PCC6803 cells via conjugation to cleave the circular chromosomes. Template DNA plasmid that has meanwhile been assimilated will then repair it achieving the desired genetic modifications. This system was validated by successfully deleting various “neutral” chromosomal loci, both individually and collectively, as well as targeting an essential gene, sll1797 . With the sacB ‐sucrose counter‐selection, all deletions were simultaneously made markerless in < 4 weeks. Moreover, we also integrate YFP with various protein degradation tags into the chromosome, allowing for their characterization at the chromosomal level. We foresee this system will greatly facilitate future genome engineering in cyanobacteria.
更新日期:2024-12-20






























 京公网安备 11010802027423号
京公网安备 11010802027423号